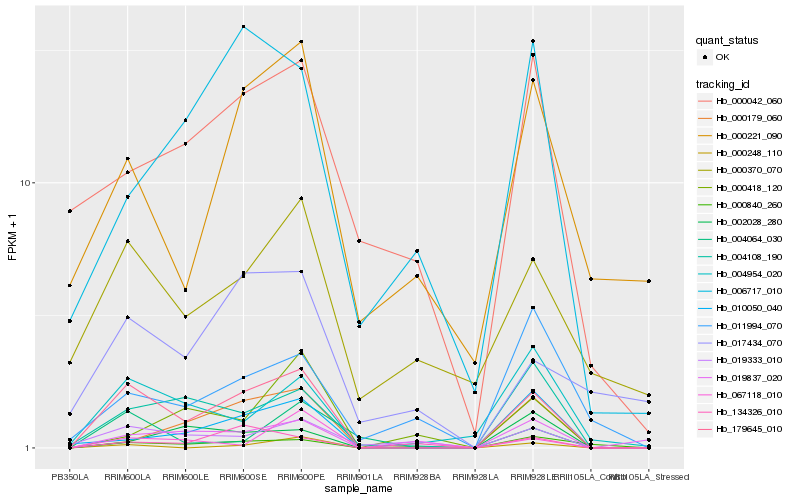
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_179645_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_004108_190 |
0.2552265945 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 3 |
Hb_019333_010 |
0.256757202 |
- |
- |
PREDICTED: uncharacterized protein LOC105633210 [Jatropha curcas] |
| 4 |
Hb_000840_260 |
0.2608008207 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0019s02540g [Populus trichocarpa] |
| 5 |
Hb_000179_060 |
0.2812763541 |
- |
- |
hypothetical protein POPTR_0010s23130g, partial [Populus trichocarpa] |
| 6 |
Hb_010050_040 |
0.2869622084 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 7 |
Hb_004954_020 |
0.2880664749 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 8 |
Hb_019837_020 |
0.3012958904 |
- |
- |
- |
| 9 |
Hb_000248_110 |
0.3014488876 |
- |
- |
hypothetical protein VITISV_037580 [Vitis vinifera] |
| 10 |
Hb_004064_030 |
0.3021877615 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 11 |
Hb_067118_010 |
0.3064253811 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 12 |
Hb_134326_010 |
0.3103782375 |
- |
- |
PREDICTED: uncharacterized protein LOC105638562 [Jatropha curcas] |
| 13 |
Hb_006717_010 |
0.3115632232 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 14 |
Hb_000370_070 |
0.3173044418 |
- |
- |
PREDICTED: subtilisin-like protease-like [Citrus sinensis] |
| 15 |
Hb_000221_090 |
0.3192356157 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 16 |
Hb_017434_070 |
0.3208146501 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_002028_280 |
0.3209977306 |
- |
- |
hypothetical protein RCOM_1397400 [Ricinus communis] |
| 18 |
Hb_000042_060 |
0.3255704418 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 19 |
Hb_000418_120 |
0.3316638702 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase [Jatropha curcas] |
| 20 |
Hb_011994_070 |
0.332543399 |
- |
- |
PREDICTED: ras-related protein RABC2a-like isoform X2 [Jatropha curcas] |