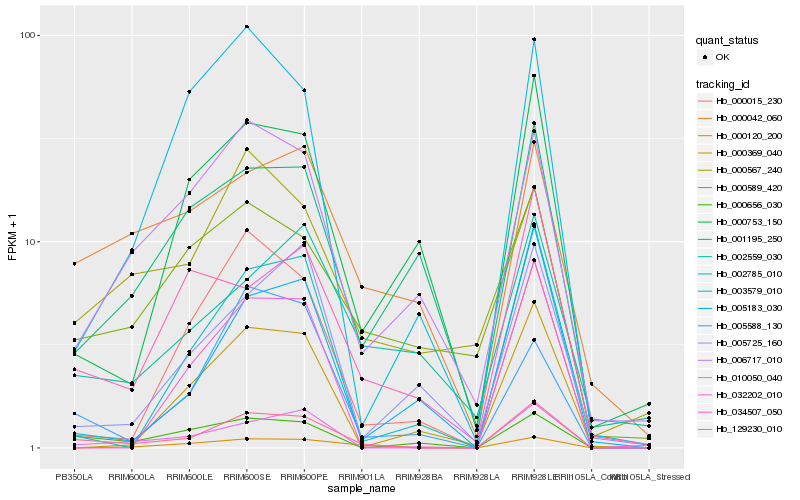
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129230_010 |
0.0 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Nicotiana sylvestris] |
| 2 |
Hb_010050_040 |
0.1647474093 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 3 |
Hb_002785_010 |
0.2110520001 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 4 |
Hb_000120_200 |
0.2129075138 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 5 |
Hb_000656_030 |
0.2150560484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_034507_050 |
0.2195640922 |
- |
- |
PREDICTED: uncharacterized protein LOC105646633 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000015_230 |
0.2250735254 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 8 |
Hb_006717_010 |
0.227300327 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 9 |
Hb_005588_130 |
0.2274796824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_032202_010 |
0.2314309678 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 11 |
Hb_000753_150 |
0.2319130628 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |
| 12 |
Hb_005183_030 |
0.2341412712 |
- |
- |
PREDICTED: probable calcium-binding protein CML16 [Jatropha curcas] |
| 13 |
Hb_000042_060 |
0.2346128311 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 14 |
Hb_005725_160 |
0.2393575001 |
- |
- |
PREDICTED: uncharacterized protein LOC105630811 [Jatropha curcas] |
| 15 |
Hb_003579_010 |
0.2396831625 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 16 |
Hb_000369_040 |
0.2397111235 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 17 |
Hb_000589_420 |
0.2402170791 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 18 |
Hb_001195_250 |
0.2426174544 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_002559_030 |
0.2438130132 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 20 |
Hb_000567_240 |
0.2441169339 |
- |
- |
PREDICTED: F-box/LRR-repeat MAX2 homolog A-like [Jatropha curcas] |