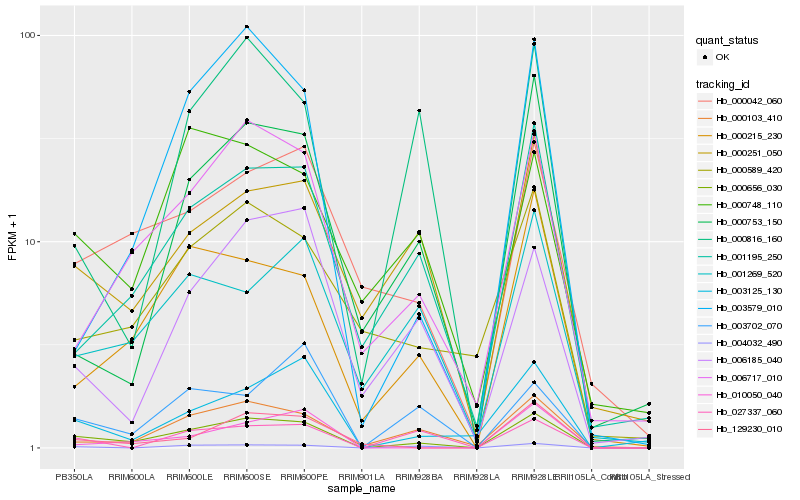
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000656_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000103_410 |
0.1840938794 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 3 |
Hb_006717_010 |
0.1845675391 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 4 |
Hb_001195_250 |
0.1950692308 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 5 |
Hb_000215_230 |
0.196166387 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 6 |
Hb_000042_060 |
0.1984832249 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 7 |
Hb_004032_490 |
0.2020710155 |
- |
- |
PREDICTED: uncharacterized protein LOC100853401 [Vitis vinifera] |
| 8 |
Hb_003125_130 |
0.2050439678 |
- |
- |
hypothetical protein JCGZ_26446 [Jatropha curcas] |
| 9 |
Hb_000251_050 |
0.2053284712 |
- |
- |
hypothetical protein JCGZ_21678 [Jatropha curcas] |
| 10 |
Hb_003579_010 |
0.2096187593 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 11 |
Hb_129230_010 |
0.2150560484 |
- |
- |
PREDICTED: inactive protein RESTRICTED TEV MOVEMENT 1-like [Nicotiana sylvestris] |
| 12 |
Hb_000589_420 |
0.2153253263 |
- |
- |
PREDICTED: uncharacterized protein LOC105647273 [Jatropha curcas] |
| 13 |
Hb_001269_520 |
0.2172536172 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 14 |
Hb_000748_110 |
0.2227156499 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 15 |
Hb_010050_040 |
0.223800525 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 16 |
Hb_027337_060 |
0.2239599924 |
- |
- |
- |
| 17 |
Hb_003702_070 |
0.226391369 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_000816_160 |
0.22658552 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 19 |
Hb_006185_040 |
0.2267346251 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |
| 20 |
Hb_000753_150 |
0.2272342682 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF5.4 [Jatropha curcas] |