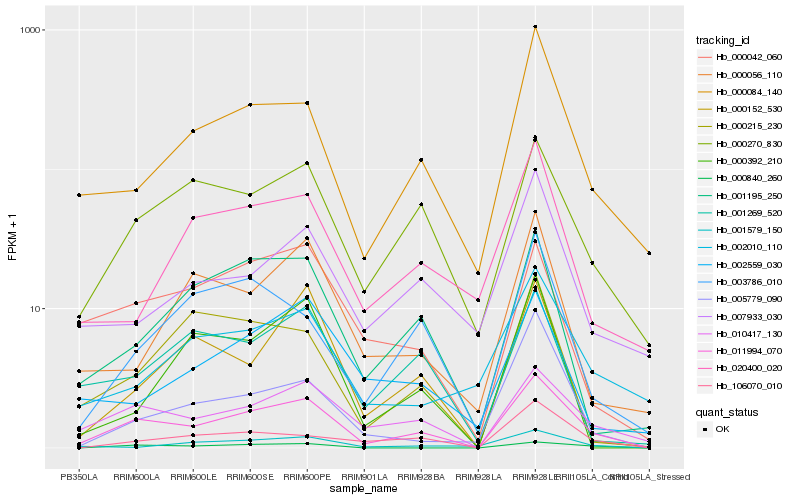
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011994_070 |
0.0 |
- |
- |
PREDICTED: ras-related protein RABC2a-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_010417_130 |
0.1534963248 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 3 |
Hb_000270_830 |
0.1799182879 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000084_140 |
0.1878329985 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 5 |
Hb_001195_250 |
0.2052636663 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 6 |
Hb_106070_010 |
0.2119744901 |
- |
- |
PREDICTED: uncharacterized protein LOC105793196 [Gossypium raimondii] |
| 7 |
Hb_000042_060 |
0.2162883156 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 8 |
Hb_001269_520 |
0.2189082207 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 9 |
Hb_003786_010 |
0.2205394576 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000840_260 |
0.2238510996 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0019s02540g [Populus trichocarpa] |
| 11 |
Hb_002010_110 |
0.2251212848 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 12 |
Hb_000215_230 |
0.2272990268 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 13 |
Hb_000152_530 |
0.2277232073 |
- |
- |
hypothetical protein SELMODRAFT_231485 [Selaginella moellendorffii] |
| 14 |
Hb_020400_020 |
0.2283618104 |
- |
- |
starch synthase isoform II [Manihot esculenta] |
| 15 |
Hb_007933_030 |
0.229258401 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 16 |
Hb_000056_110 |
0.2293943581 |
- |
- |
PREDICTED: uncharacterized protein LOC105630405 [Jatropha curcas] |
| 17 |
Hb_001579_150 |
0.2316137332 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_002559_030 |
0.231809158 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 19 |
Hb_005779_090 |
0.2321347153 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 20 |
Hb_000392_210 |
0.2357687199 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0004s04600g [Populus trichocarpa] |