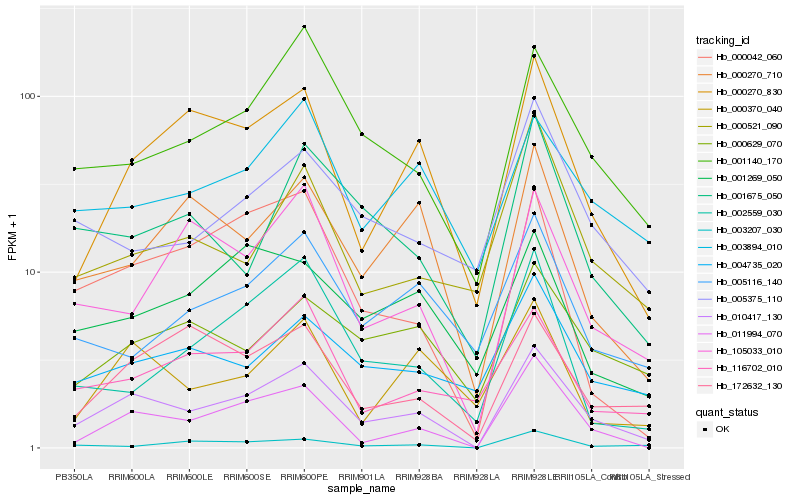
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010417_130 |
0.0 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 2 |
Hb_011994_070 |
0.1534963248 |
- |
- |
PREDICTED: ras-related protein RABC2a-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001140_170 |
0.1657623421 |
- |
- |
PREDICTED: remorin [Jatropha curcas] |
| 4 |
Hb_000270_830 |
0.1700923771 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_000042_060 |
0.1825537668 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 6 |
Hb_105033_010 |
0.1869457534 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000270_710 |
0.1871840757 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003894_010 |
0.1912479415 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_172632_130 |
0.1920782232 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 10 |
Hb_001675_050 |
0.1924789385 |
- |
- |
PREDICTED: uncharacterized protein LOC105635000 [Jatropha curcas] |
| 11 |
Hb_004735_020 |
0.1935662325 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 12 |
Hb_000629_070 |
0.1976664831 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 13 |
Hb_116702_010 |
0.1981843174 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 14 |
Hb_000370_040 |
0.198724415 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 15 |
Hb_005375_110 |
0.1996879782 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 16 |
Hb_000521_090 |
0.1999264576 |
- |
- |
PREDICTED: RING-H2 finger protein ATL5 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005116_140 |
0.2034857846 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 18 |
Hb_002559_030 |
0.2044351942 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 19 |
Hb_003207_030 |
0.2047755869 |
- |
- |
hypothetical protein VITISV_027754 [Vitis vinifera] |
| 20 |
Hb_001269_050 |
0.2059070109 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |