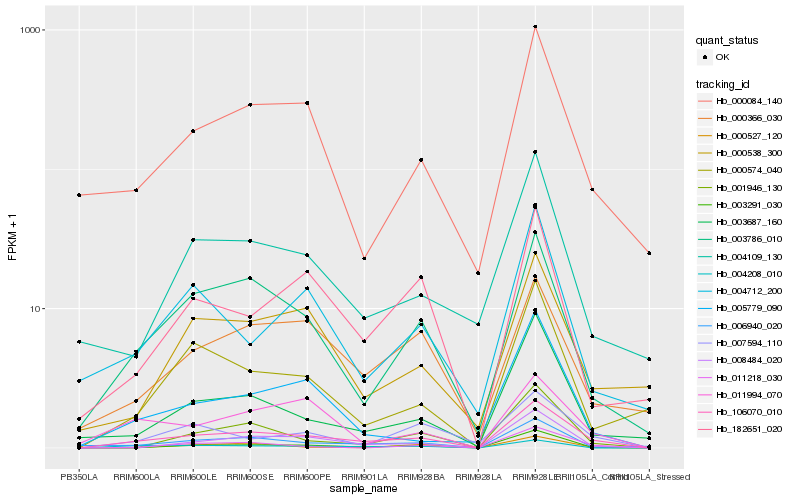
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106070_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105793196 [Gossypium raimondii] |
| 2 |
Hb_003291_030 |
0.1770730229 |
- |
- |
Gag-pol protein, putative [Solanum demissum] |
| 3 |
Hb_003786_010 |
0.1794244358 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_008484_020 |
0.197059104 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 5 |
Hb_000084_140 |
0.2009983918 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 6 |
Hb_006940_020 |
0.206055148 |
- |
- |
polyprotein [Oryza australiensis] |
| 7 |
Hb_004208_010 |
0.2075227517 |
- |
- |
PREDICTED: uncharacterized protein LOC105914588 [Setaria italica] |
| 8 |
Hb_000366_030 |
0.2076087847 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Jatropha curcas] |
| 9 |
Hb_003687_160 |
0.2092258284 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 10 |
Hb_001946_130 |
0.2112885525 |
- |
- |
- |
| 11 |
Hb_182651_020 |
0.2116874963 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_011994_070 |
0.2119744901 |
- |
- |
PREDICTED: ras-related protein RABC2a-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_011218_030 |
0.2122016986 |
- |
- |
hypothetical protein VITISV_002891 [Vitis vinifera] |
| 14 |
Hb_007594_110 |
0.2129276855 |
- |
- |
fructokinase [Manihot esculenta] |
| 15 |
Hb_004109_130 |
0.2152516346 |
- |
- |
PREDICTED: translation initiation factor IF-2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005779_090 |
0.2182299753 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 17 |
Hb_000538_300 |
0.218518611 |
- |
- |
PREDICTED: WAT1-related protein At3g02690, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000574_040 |
0.2195998166 |
- |
- |
PREDICTED: triphosphate tunel metalloenzyme 3-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000527_120 |
0.2226317777 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 20 |
Hb_004712_200 |
0.2228879688 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |