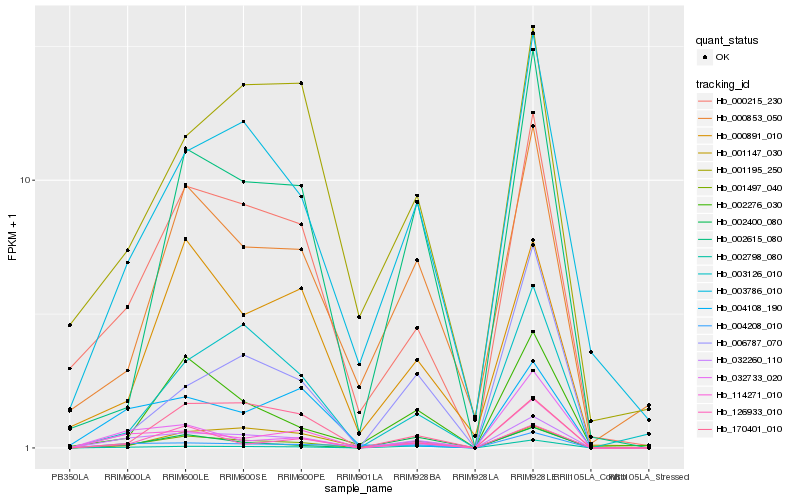
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032260_110 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001497_040 |
0.1073236585 |
- |
- |
hypothetical protein POPTR_0008s219701g, partial [Populus trichocarpa] |
| 3 |
Hb_114271_010 |
0.1498843769 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 4 |
Hb_000215_230 |
0.1732931466 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 5 |
Hb_003786_010 |
0.1790854937 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002798_080 |
0.1897650372 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 7 |
Hb_004208_010 |
0.1913661666 |
- |
- |
PREDICTED: uncharacterized protein LOC105914588 [Setaria italica] |
| 8 |
Hb_002400_080 |
0.2044419942 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 9 |
Hb_002615_080 |
0.216926007 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 10 |
Hb_003126_010 |
0.2203330768 |
- |
- |
PREDICTED: oligopeptide transporter 7-like isoform X1 [Malus domestica] |
| 11 |
Hb_126933_010 |
0.2204421211 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 12 |
Hb_000853_050 |
0.2213611684 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_001147_030 |
0.2222161198 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 14 |
Hb_170401_010 |
0.2230559919 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 15 |
Hb_001195_250 |
0.2259228035 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 16 |
Hb_004108_190 |
0.2292483316 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 17 |
Hb_006787_070 |
0.2307446264 |
- |
- |
PREDICTED: uncharacterized protein LOC104121706, partial [Nicotiana tomentosiformis] |
| 18 |
Hb_000891_010 |
0.2339807805 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002276_030 |
0.2341597182 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 20 |
Hb_032733_020 |
0.2355188224 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X4 [Citrus sinensis] |