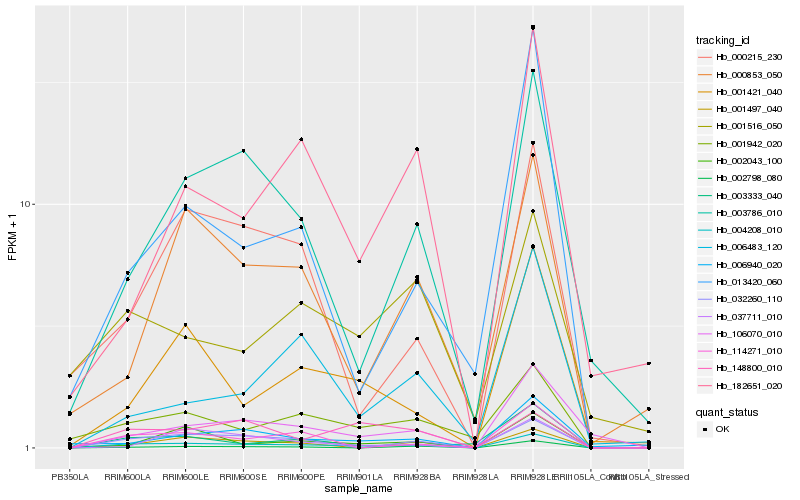
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004208_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105914588 [Setaria italica] |
| 2 |
Hb_032260_110 |
0.1913661666 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001942_020 |
0.1979546602 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_106070_010 |
0.2075227517 |
- |
- |
PREDICTED: uncharacterized protein LOC105793196 [Gossypium raimondii] |
| 5 |
Hb_001421_040 |
0.2093229237 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 2 [Jatropha curcas] |
| 6 |
Hb_003786_010 |
0.211622012 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_114271_010 |
0.2156740968 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 8 |
Hb_148800_010 |
0.223092749 |
- |
- |
- |
| 9 |
Hb_001497_040 |
0.2232747848 |
- |
- |
hypothetical protein POPTR_0008s219701g, partial [Populus trichocarpa] |
| 10 |
Hb_002798_080 |
0.2289981947 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 11 |
Hb_006940_020 |
0.2334836773 |
- |
- |
polyprotein [Oryza australiensis] |
| 12 |
Hb_000215_230 |
0.234550668 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 13 |
Hb_002043_100 |
0.2345606221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_182651_020 |
0.2368329613 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_013420_060 |
0.2378537651 |
- |
- |
NADH dehydrogenase subunit 7 [Nymphaea sp. Bergthorsson 0401] |
| 16 |
Hb_003333_040 |
0.2405168852 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000853_050 |
0.2415417436 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_001516_050 |
0.2451897991 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 19 |
Hb_037711_010 |
0.2473980726 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 20 |
Hb_006483_120 |
0.248794149 |
- |
- |
DNA binding protein, putative [Ricinus communis] |