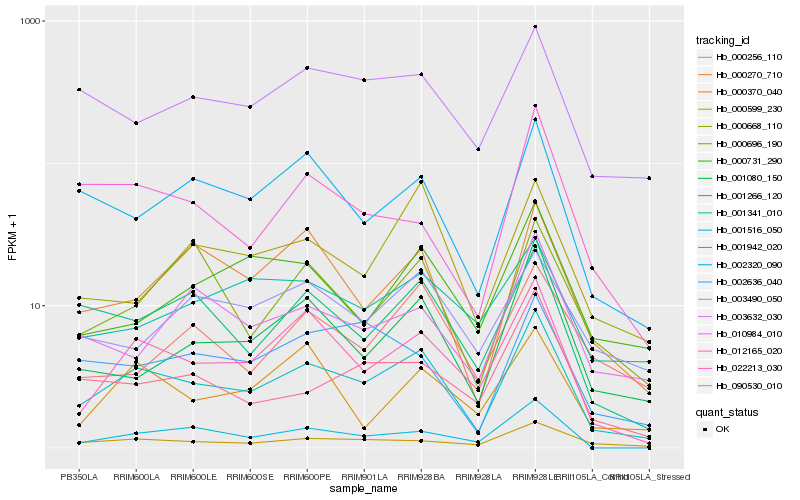
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001516_050 |
0.0 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 2 |
Hb_022213_030 |
0.1327489378 |
- |
- |
PREDICTED: uncharacterized protein LOC105114631 [Populus euphratica] |
| 3 |
Hb_000599_230 |
0.1571281414 |
- |
- |
- |
| 4 |
Hb_001266_120 |
0.1578015173 |
- |
- |
auxilin, putative [Ricinus communis] |
| 5 |
Hb_002320_090 |
0.1582212757 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 6 |
Hb_001080_150 |
0.160731104 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 7 |
Hb_001942_020 |
0.1627353733 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000270_710 |
0.1652630907 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_003632_030 |
0.1772210431 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 10 |
Hb_002636_040 |
0.1830401429 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000668_110 |
0.1843147016 |
- |
- |
PREDICTED: acetyl-CoA carboxylase 1-like [Jatropha curcas] |
| 12 |
Hb_010984_010 |
0.1845899249 |
- |
- |
PREDICTED: translation factor GUF1 homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_090530_010 |
0.1854328085 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 14 |
Hb_012165_020 |
0.1856389659 |
- |
- |
hypothetical protein POPTR_0007s05770g [Populus trichocarpa] |
| 15 |
Hb_000696_190 |
0.1886460215 |
- |
- |
PREDICTED: protein MID1-COMPLEMENTING ACTIVITY 1 [Jatropha curcas] |
| 16 |
Hb_000731_290 |
0.1901005161 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_003490_050 |
0.1928284925 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000256_110 |
0.192839161 |
- |
- |
PREDICTED: aspartate aminotransferase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000370_040 |
0.1938568416 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 20 |
Hb_001341_010 |
0.1950200858 |
- |
- |
PREDICTED: carbamoyl-phosphate synthase large chain, chloroplastic [Jatropha curcas] |