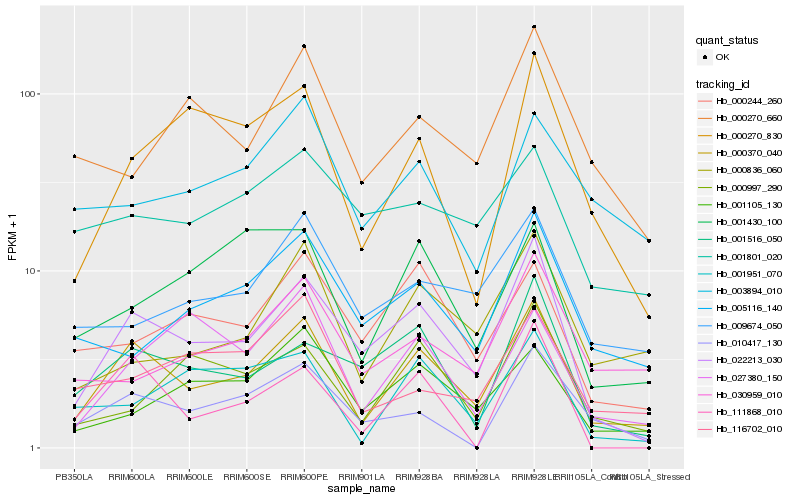
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000370_040 |
0.0 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 2 |
Hb_022213_030 |
0.1293961021 |
- |
- |
PREDICTED: uncharacterized protein LOC105114631 [Populus euphratica] |
| 3 |
Hb_000270_830 |
0.192076736 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000836_060 |
0.1931404479 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-2 [Jatropha curcas] |
| 5 |
Hb_001516_050 |
0.1938568416 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 6 |
Hb_010417_130 |
0.198724415 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 7 |
Hb_009674_050 |
0.2028775939 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 8 |
Hb_000244_260 |
0.2040237646 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001105_130 |
0.2071230341 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 10 |
Hb_116702_010 |
0.2087426311 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 11 |
Hb_030959_010 |
0.2088453861 |
- |
- |
cullin, putative [Ricinus communis] |
| 12 |
Hb_005116_140 |
0.2089742755 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 13 |
Hb_001430_100 |
0.209767795 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_001951_070 |
0.2107813406 |
- |
- |
putative protein [Arabidopsis thaliana] |
| 15 |
Hb_027380_150 |
0.2120689189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003894_010 |
0.2121774051 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000997_290 |
0.2124036789 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 18 |
Hb_111868_010 |
0.2137191542 |
- |
- |
hypothetical protein CICLE_v10029531mg [Citrus clementina] |
| 19 |
Hb_001801_020 |
0.2146492656 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 20 |
Hb_000270_660 |
0.2157685639 |
- |
- |
purine permease, putative [Ricinus communis] |