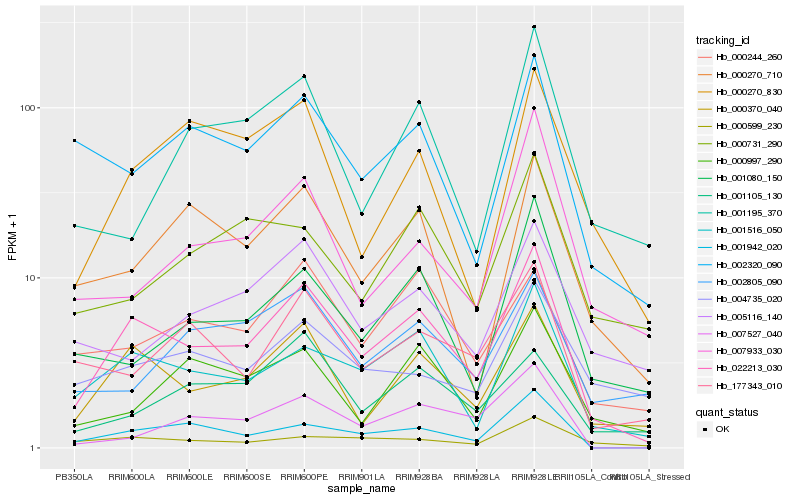
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022213_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105114631 [Populus euphratica] |
| 2 |
Hb_000370_040 |
0.1293961021 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 3 |
Hb_001516_050 |
0.1327489378 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD3-1 [Jatropha curcas] |
| 4 |
Hb_001942_020 |
0.1548512698 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001080_150 |
0.1872852169 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 6 |
Hb_177343_010 |
0.1875323599 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 7 |
Hb_000270_710 |
0.1883273607 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002320_090 |
0.1916376009 |
- |
- |
PREDICTED: endoglucanase 25-like [Jatropha curcas] |
| 9 |
Hb_000270_830 |
0.1918954589 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000244_260 |
0.1931155651 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007527_040 |
0.194415721 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 12 |
Hb_000599_230 |
0.1979303375 |
- |
- |
- |
| 13 |
Hb_000997_290 |
0.19829405 |
- |
- |
PREDICTED: acylamino-acid-releasing enzyme [Jatropha curcas] |
| 14 |
Hb_005116_140 |
0.201377345 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |
| 15 |
Hb_002805_090 |
0.203720964 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 16 |
Hb_007933_030 |
0.2040152392 |
- |
- |
PREDICTED: uncharacterized protein LOC105639932 [Jatropha curcas] |
| 17 |
Hb_000731_290 |
0.2049969576 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 18 |
Hb_001105_130 |
0.206384649 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 19 |
Hb_001195_370 |
0.2069101004 |
- |
- |
- |
| 20 |
Hb_004735_020 |
0.2070146307 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |