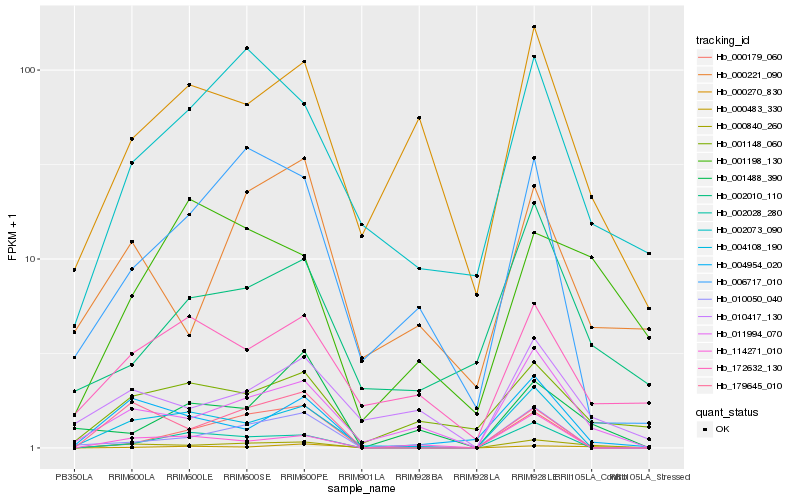
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000840_260 |
0.0 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0019s02540g [Populus trichocarpa] |
| 2 |
Hb_011994_070 |
0.2238510996 |
- |
- |
PREDICTED: ras-related protein RABC2a-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_179645_010 |
0.2608008207 |
- |
- |
- |
| 4 |
Hb_004954_020 |
0.2613695181 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 5 |
Hb_004108_190 |
0.2658973156 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 6 |
Hb_000483_330 |
0.2809160044 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH6-like [Jatropha curcas] |
| 7 |
Hb_002028_280 |
0.287680879 |
- |
- |
hypothetical protein RCOM_1397400 [Ricinus communis] |
| 8 |
Hb_002073_090 |
0.2901606875 |
- |
- |
PREDICTED: copper transporter 6-like [Jatropha curcas] |
| 9 |
Hb_010050_040 |
0.2909918829 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 10 |
Hb_001148_060 |
0.292496571 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 11 |
Hb_010417_130 |
0.2983781921 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 2 [Jatropha curcas] |
| 12 |
Hb_000179_060 |
0.3047501707 |
- |
- |
hypothetical protein POPTR_0010s23130g, partial [Populus trichocarpa] |
| 13 |
Hb_002010_110 |
0.3103964306 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 14 |
Hb_000221_090 |
0.3114312617 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-6-like [Jatropha curcas] |
| 15 |
Hb_000270_830 |
0.3140543105 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_006717_010 |
0.3143107587 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 17 |
Hb_114271_010 |
0.3143792927 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 18 |
Hb_001198_130 |
0.3174715138 |
- |
- |
PREDICTED: potassium channel AKT2/3 [Jatropha curcas] |
| 19 |
Hb_001488_390 |
0.317651721 |
- |
- |
SP1L, putative [Ricinus communis] |
| 20 |
Hb_172632_130 |
0.3176922827 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |