| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
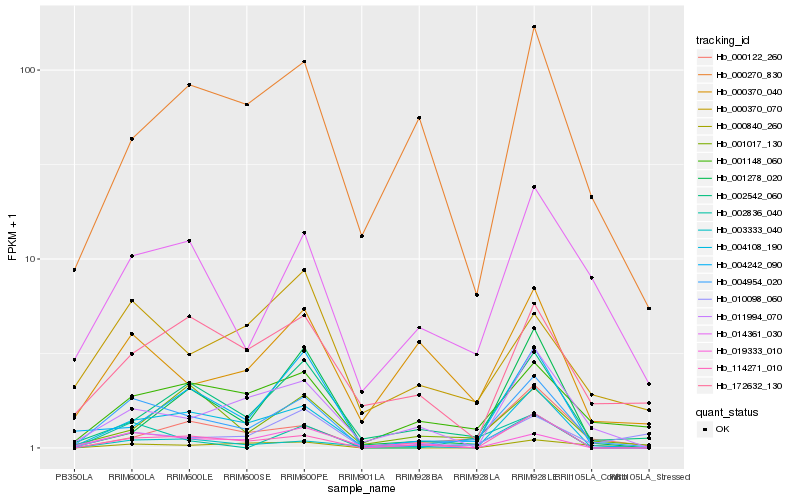
Hb_004954_020 |
0.0 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004108_190 |
0.2120198476 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 3 |
Hb_114271_010 |
0.2346085828 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 4 |
Hb_001148_060 |
0.2386221993 |
- |
- |
hypothetical protein JCGZ_00278 [Jatropha curcas] |
| 5 |
Hb_014361_030 |
0.2386634335 |
- |
- |
hypothetical protein POPTR_0005s18860g [Populus trichocarpa] |
| 6 |
Hb_002542_060 |
0.2431333072 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 7 |
Hb_011994_070 |
0.2513485455 |
- |
- |
PREDICTED: ras-related protein RABC2a-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000122_260 |
0.2599464698 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 1-like [Jatropha curcas] |
| 9 |
Hb_004242_090 |
0.2602554577 |
- |
- |
hypothetical protein JCGZ_00767 [Jatropha curcas] |
| 10 |
Hb_000840_260 |
0.2613695181 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0019s02540g [Populus trichocarpa] |
| 11 |
Hb_000370_040 |
0.2662033387 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 12 |
Hb_002836_040 |
0.2683861617 |
- |
- |
Auxin-induced protein 5NG4 [Glycine soja] |
| 13 |
Hb_010098_060 |
0.268671867 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g17580 [Jatropha curcas] |
| 14 |
Hb_001017_130 |
0.2708325079 |
- |
- |
hypothetical protein POPTR_0010s06070g [Populus trichocarpa] |
| 15 |
Hb_000370_070 |
0.2744802955 |
- |
- |
PREDICTED: subtilisin-like protease-like [Citrus sinensis] |
| 16 |
Hb_003333_040 |
0.2744816443 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_019333_010 |
0.2766889887 |
- |
- |
PREDICTED: uncharacterized protein LOC105633210 [Jatropha curcas] |
| 18 |
Hb_000270_830 |
0.2782120271 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_172632_130 |
0.2798376769 |
- |
- |
hypothetical protein JCGZ_22584 [Jatropha curcas] |
| 20 |
Hb_001278_020 |
0.2845083846 |
- |
- |
sucrose synthase 6 [Hevea brasiliensis] |