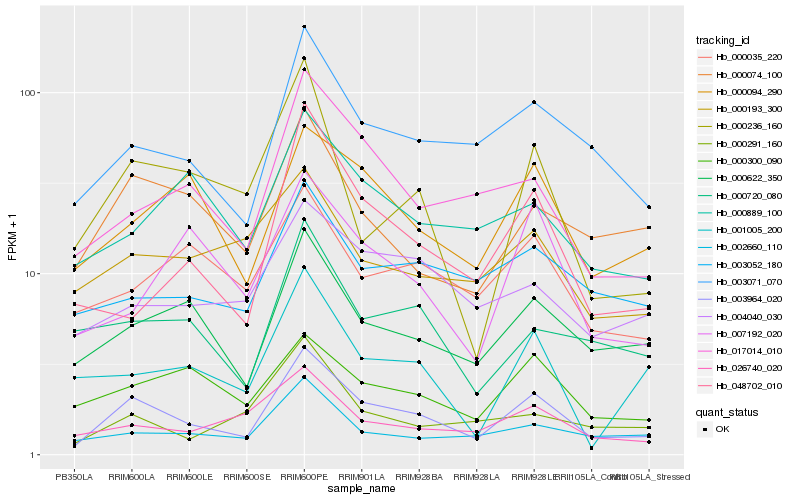
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003964_020 |
0.0 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1621120 [Ricinus communis] |
| 2 |
Hb_003071_070 |
0.1580103348 |
- |
- |
sucrose synthase 3 [Hevea brasiliensis] |
| 3 |
Hb_000622_350 |
0.1659637536 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 4 |
Hb_017014_010 |
0.1775163792 |
- |
- |
PREDICTED: syntaxin-132-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000074_100 |
0.1792229225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000094_290 |
0.1805166544 |
- |
- |
PREDICTED: outer envelope pore protein 24B, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_002660_110 |
0.1851683507 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein CCR1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_026740_020 |
0.1895254672 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 9 |
Hb_000300_090 |
0.1909153414 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_000236_160 |
0.1924357552 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 11 |
Hb_000720_080 |
0.1926440747 |
- |
- |
PREDICTED: bifunctional UDP-glucose 4-epimerase and UDP-xylose 4-epimerase 1 [Jatropha curcas] |
| 12 |
Hb_048702_010 |
0.1930618706 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_003052_180 |
0.1937726716 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 14 |
Hb_004040_030 |
0.1971234646 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000889_100 |
0.1989791066 |
- |
- |
PREDICTED: syntaxin-132-like isoform X2 [Sesamum indicum] |
| 16 |
Hb_000035_220 |
0.1992084991 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 8 [Jatropha curcas] |
| 17 |
Hb_000291_160 |
0.2004482459 |
- |
- |
PREDICTED: uncharacterized protein LOC105638863 [Jatropha curcas] |
| 18 |
Hb_007192_020 |
0.2030790831 |
- |
- |
PREDICTED: high-affinity nitrate transporter 3.1-like [Jatropha curcas] |
| 19 |
Hb_001005_200 |
0.2034488593 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000193_300 |
0.2039532275 |
- |
- |
PREDICTED: putative glycosyltransferase 5 [Jatropha curcas] |