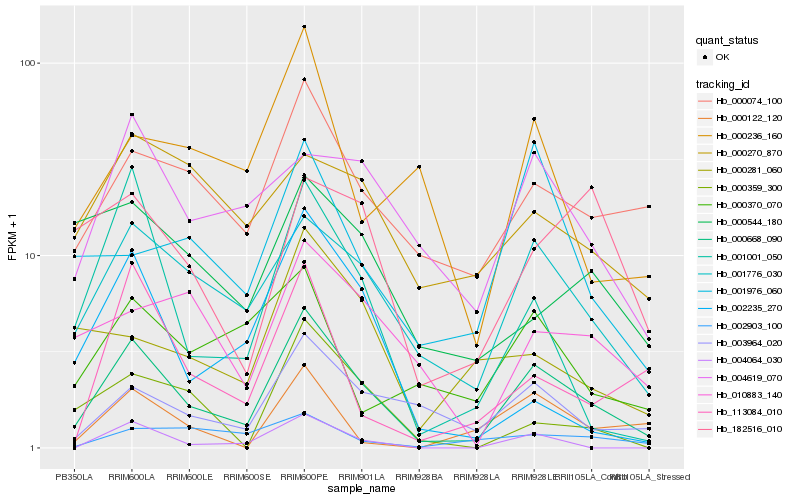
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000668_090 |
0.0 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 2 |
Hb_001776_030 |
0.1824391831 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003964_020 |
0.2178229602 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1621120 [Ricinus communis] |
| 4 |
Hb_000074_100 |
0.222040324 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001001_050 |
0.2371779807 |
- |
- |
CXE protein [Hevea brasiliensis] |
| 6 |
Hb_004619_070 |
0.245711223 |
- |
- |
hypothetical protein POPTR_0006s00550g [Populus trichocarpa] |
| 7 |
Hb_004064_030 |
0.2506409436 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 8 |
Hb_010883_140 |
0.2572566919 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_113084_010 |
0.2647670836 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 10 |
Hb_000236_160 |
0.2650866476 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 11 |
Hb_000359_300 |
0.2657501456 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At1g06470 [Zea mays] |
| 12 |
Hb_000370_070 |
0.2671533802 |
- |
- |
PREDICTED: subtilisin-like protease-like [Citrus sinensis] |
| 13 |
Hb_001976_060 |
0.2685911553 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000122_120 |
0.2703650644 |
- |
- |
PREDICTED: arogenate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000281_060 |
0.2741334901 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_002903_100 |
0.2742166018 |
- |
- |
hypothetical protein POPTR_0011s04750g [Populus trichocarpa] |
| 17 |
Hb_182516_010 |
0.2754002647 |
- |
- |
PREDICTED: uncharacterized protein LOC105641595 [Jatropha curcas] |
| 18 |
Hb_000270_870 |
0.2755144828 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like [Jatropha curcas] |
| 19 |
Hb_000544_180 |
0.2767678957 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X1 [Jatropha curcas] |
| 20 |
Hb_002235_270 |
0.2773374162 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100 [Jatropha curcas] |