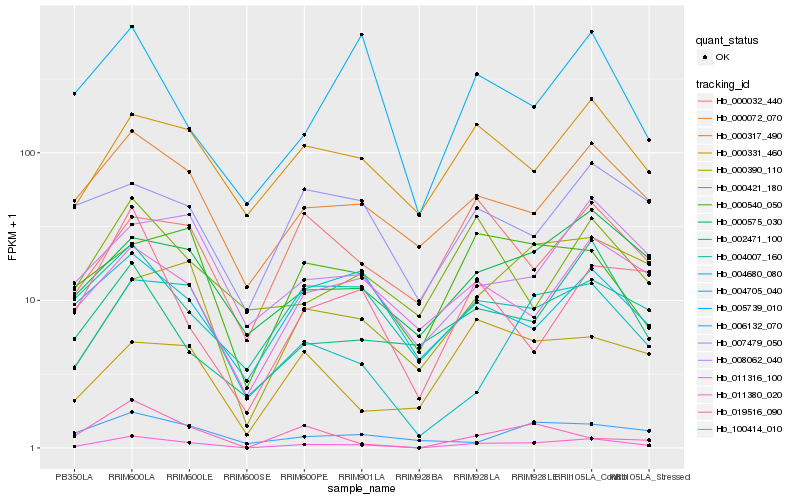
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011380_020 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X4 [Populus euphratica] |
| 2 |
Hb_000072_070 |
0.2055774269 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl diphosphate synthase [Hevea brasiliensis] |
| 3 |
Hb_004680_080 |
0.2120640894 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 4 |
Hb_000575_030 |
0.2222188646 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_000317_490 |
0.2242100508 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 6 |
Hb_011316_100 |
0.2317700445 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4-like [Jatropha curcas] |
| 7 |
Hb_008062_040 |
0.2359342875 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000540_050 |
0.2399329494 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 9 |
Hb_004007_160 |
0.2406503264 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 10 |
Hb_002471_100 |
0.2439055925 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_100414_010 |
0.2444167223 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl pyrophosphate synthase [Hevea brasiliensis] |
| 12 |
Hb_000390_110 |
0.247167471 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 13 |
Hb_004705_040 |
0.2473276515 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 14 |
Hb_000421_180 |
0.2537454068 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 15 |
Hb_000032_440 |
0.2537462901 |
- |
- |
protein with unknown function [Ricinus communis] |
| 16 |
Hb_006132_070 |
0.2546985784 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 17 |
Hb_005739_010 |
0.2548394005 |
- |
- |
hypothetical protein JCGZ_04016 [Jatropha curcas] |
| 18 |
Hb_019516_090 |
0.2548458452 |
- |
- |
hypothetical protein POPTR_0004s06350g [Populus trichocarpa] |
| 19 |
Hb_007479_050 |
0.2555409974 |
- |
- |
PREDICTED: uncharacterized protein LOC105649412 [Jatropha curcas] |
| 20 |
Hb_000331_460 |
0.2563190282 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |