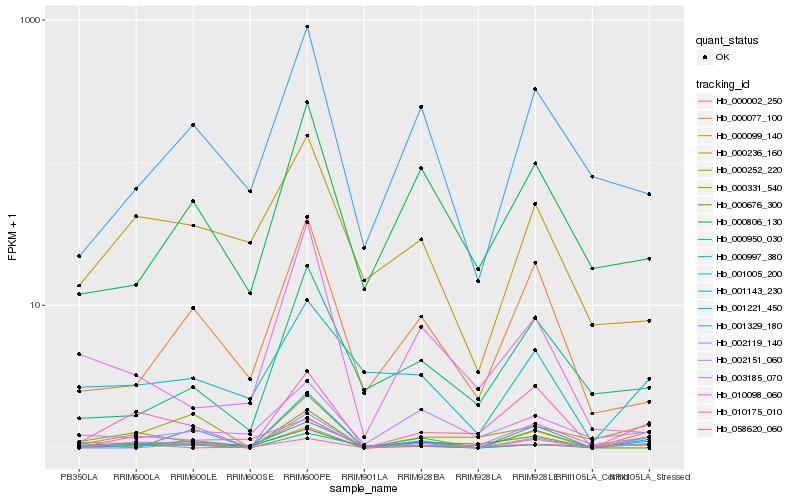
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_540 |
0.0 |
- |
- |
hypothetical protein POPTR_0015s10070g, partial [Populus trichocarpa] |
| 2 |
Hb_010175_010 |
0.2145998954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000002_250 |
0.2283482864 |
- |
- |
hypothetical protein JCGZ_02310 [Jatropha curcas] |
| 4 |
Hb_001143_230 |
0.2637630889 |
- |
- |
beta-galactosidase family protein [Populus trichocarpa] |
| 5 |
Hb_000997_380 |
0.2928669507 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 6 |
Hb_001005_200 |
0.296969311 |
- |
- |
PREDICTED: receptor-like protein kinase HERK 1 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000236_160 |
0.3117521329 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 6 [Jatropha curcas] |
| 8 |
Hb_000099_140 |
0.3184743777 |
- |
- |
PREDICTED: aluminum-activated malate transporter 9-like [Jatropha curcas] |
| 9 |
Hb_001329_180 |
0.3228107915 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 10 |
Hb_002151_060 |
0.3232244768 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc4 [Jatropha curcas] |
| 11 |
Hb_001221_450 |
0.3236277764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000252_220 |
0.3276378704 |
- |
- |
PREDICTED: copper amine oxidase 1-like isoform X2 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_003185_070 |
0.3301901546 |
- |
- |
Plant natriuretic peptide A [Theobroma cacao] |
| 14 |
Hb_058620_060 |
0.3322568897 |
- |
- |
uncharacterized protein LOC105646159 precursor [Jatropha curcas] |
| 15 |
Hb_000806_130 |
0.3332622873 |
- |
- |
Adenosine kinase 2 [Theobroma cacao] |
| 16 |
Hb_000950_030 |
0.3346021938 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 17 |
Hb_000077_100 |
0.3354866715 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002119_140 |
0.3368523762 |
- |
- |
PREDICTED: uncharacterized protein LOC105634172 [Jatropha curcas] |
| 19 |
Hb_000676_300 |
0.3381946268 |
- |
- |
PREDICTED: probable caffeine synthase 4 [Jatropha curcas] |
| 20 |
Hb_010098_060 |
0.3383024394 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g17580 [Jatropha curcas] |