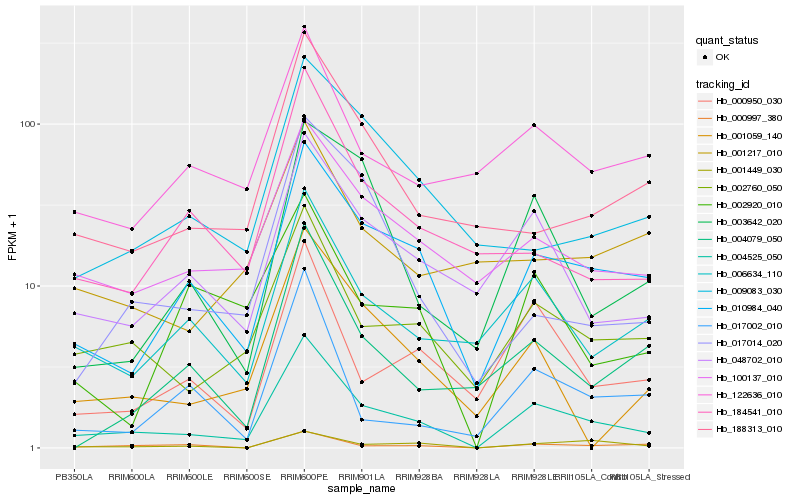
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004525_050 |
0.0 |
- |
- |
similar to Drosophila melanogaster CG10960, partial [Drosophila yakuba] |
| 2 |
Hb_010984_040 |
0.151252423 |
- |
- |
hypothetical protein M569_17286, partial [Genlisea aurea] |
| 3 |
Hb_002760_050 |
0.1637803164 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_048702_010 |
0.1847475471 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_100137_010 |
0.1923953238 |
- |
- |
hypothetical protein JCGZ_17848 [Jatropha curcas] |
| 6 |
Hb_006634_110 |
0.1927493161 |
- |
- |
BnaC04g40780D [Brassica napus] |
| 7 |
Hb_000950_030 |
0.2018696129 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g10290 [Nelumbo nucifera] |
| 8 |
Hb_000997_380 |
0.2040977986 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 9 |
Hb_001449_030 |
0.2085364126 |
- |
- |
PREDICTED: RNA polymerase II-associated protein 3 isoform X3 [Jatropha curcas] |
| 10 |
Hb_001059_140 |
0.2127624298 |
- |
- |
hypothetical protein POPTR_0004s03390g [Populus trichocarpa] |
| 11 |
Hb_188313_010 |
0.2187315392 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 12 |
Hb_122636_010 |
0.2188922 |
- |
- |
dienelactone hydrolase family protein [Populus trichocarpa] |
| 13 |
Hb_184541_010 |
0.2221943588 |
- |
- |
PREDICTED: selT-like protein [Jatropha curcas] |
| 14 |
Hb_003642_020 |
0.2276430004 |
- |
- |
chlorophyll(ide) b reductase, partial [Pyrus x bretschneideri] |
| 15 |
Hb_004079_050 |
0.2282339714 |
- |
- |
hypothetical protein JCGZ_07229 [Jatropha curcas] |
| 16 |
Hb_017002_010 |
0.2286311646 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 17 |
Hb_002920_010 |
0.232494031 |
- |
- |
PREDICTED: sulfiredoxin, chloroplastic/mitochondrial [Populus euphratica] |
| 18 |
Hb_009083_030 |
0.2337352764 |
- |
- |
hypothetical protein POPTR_0007s14760g [Populus trichocarpa] |
| 19 |
Hb_017014_020 |
0.2338334506 |
- |
- |
- |
| 20 |
Hb_001217_010 |
0.2391297784 |
- |
- |
hypothetical protein JCGZ_04022 [Jatropha curcas] |