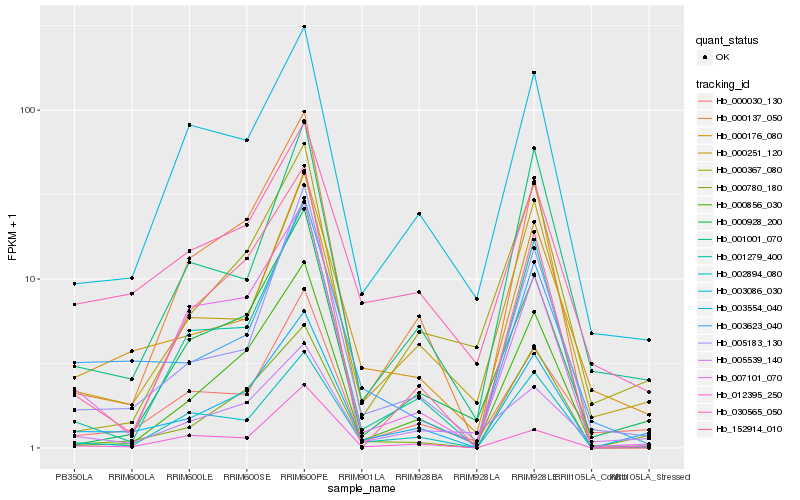
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003554_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000780_180 |
0.1262547807 |
- |
- |
PREDICTED: uncharacterized protein LOC105641547 [Jatropha curcas] |
| 3 |
Hb_000176_080 |
0.1429664215 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g30520 [Jatropha curcas] |
| 4 |
Hb_000030_130 |
0.1432045194 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000137_050 |
0.1510450052 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002894_080 |
0.1545642221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005183_130 |
0.1550404393 |
- |
- |
glycogenin, putative [Ricinus communis] |
| 8 |
Hb_000251_120 |
0.1552315291 |
- |
- |
O-acetyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000856_030 |
0.16119941 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_003086_030 |
0.1635585278 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 17 [Jatropha curcas] |
| 11 |
Hb_030565_050 |
0.1640503434 |
- |
- |
UDP-D-GLUCURONATE 4-EPIMERASE 6 family protein [Populus trichocarpa] |
| 12 |
Hb_003623_040 |
0.1643906545 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 13 |
Hb_000928_200 |
0.1649411335 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 14 |
Hb_001001_070 |
0.1653125468 |
- |
- |
PREDICTED: uncharacterized protein LOC105633663 [Jatropha curcas] |
| 15 |
Hb_001279_400 |
0.1694804401 |
- |
- |
PREDICTED: uncharacterized protein LOC105633544 [Jatropha curcas] |
| 16 |
Hb_152914_010 |
0.1704307728 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 17 |
Hb_005539_140 |
0.1727934306 |
- |
- |
PREDICTED: U-box domain-containing protein 19-like [Jatropha curcas] |
| 18 |
Hb_012395_250 |
0.1750426181 |
- |
- |
hypothetical protein RCOM_1217340 [Ricinus communis] |
| 19 |
Hb_000367_080 |
0.1769891694 |
- |
- |
PREDICTED: UDP-glucose 6-dehydrogenase 1-like [Jatropha curcas] |
| 20 |
Hb_007101_070 |
0.1784365365 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1 [Jatropha curcas] |