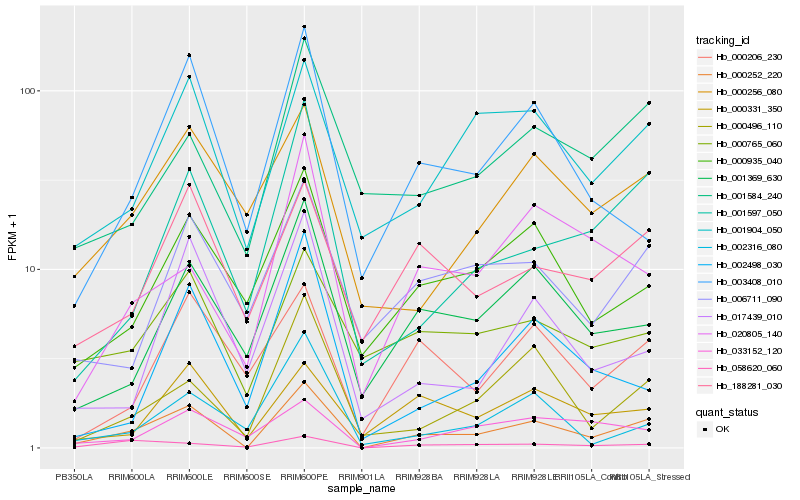
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000252_220 |
0.0 |
- |
- |
PREDICTED: copper amine oxidase 1-like isoform X2 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_001904_050 |
0.2144342378 |
- |
- |
alpha-tubulin 1 [Hevea brasiliensis] |
| 3 |
Hb_001597_050 |
0.2159930627 |
- |
- |
- |
| 4 |
Hb_000496_110 |
0.2166064001 |
- |
- |
PREDICTED: uncharacterized protein LOC105650214 [Jatropha curcas] |
| 5 |
Hb_001369_630 |
0.2200584971 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 3-like [Jatropha curcas] |
| 6 |
Hb_058620_060 |
0.2222499023 |
- |
- |
uncharacterized protein LOC105646159 precursor [Jatropha curcas] |
| 7 |
Hb_000331_350 |
0.2258088221 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 8 |
Hb_000935_040 |
0.2259661595 |
- |
- |
Uncharacterized protein isoform 2 [Theobroma cacao] |
| 9 |
Hb_188281_030 |
0.2277271213 |
- |
- |
PREDICTED: uridine nucleosidase 1 [Jatropha curcas] |
| 10 |
Hb_003408_010 |
0.229665036 |
- |
- |
fructokinase [Manihot esculenta] |
| 11 |
Hb_001584_240 |
0.232526097 |
- |
- |
ADP-ribosylation factor [Gossypium barbadense] |
| 12 |
Hb_006711_090 |
0.2328028694 |
- |
- |
PREDICTED: fatty acid 2-hydroxylase 1-like [Jatropha curcas] |
| 13 |
Hb_000765_060 |
0.2335161422 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 14 |
Hb_000256_080 |
0.2355331748 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 15 |
Hb_000206_230 |
0.236168197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_033152_120 |
0.2412532563 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 17 |
Hb_017439_010 |
0.2424250426 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 18 |
Hb_020805_140 |
0.2440307246 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g42360-like [Jatropha curcas] |
| 19 |
Hb_002498_030 |
0.2452207952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002316_080 |
0.2456871956 |
- |
- |
PREDICTED: RING-H2 finger protein ATL46-like [Populus euphratica] |