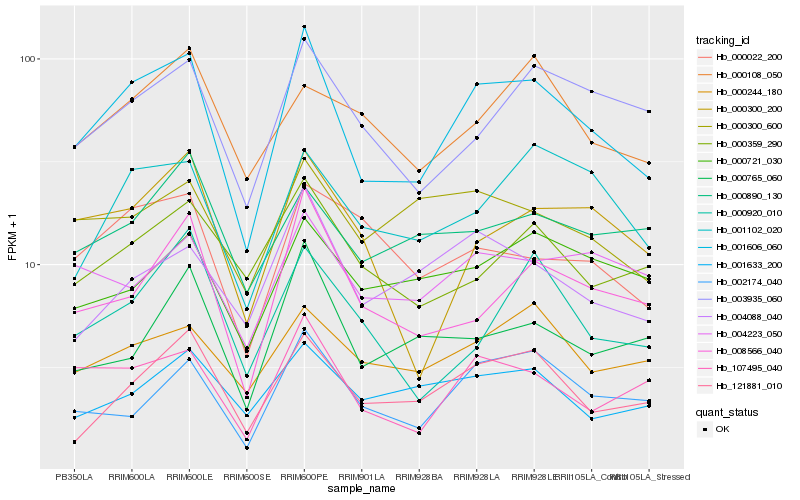
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001606_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_107495_040 |
0.128840726 |
- |
- |
PREDICTED: 12-oxophytodienoate reductase 2-like [Citrus sinensis] |
| 3 |
Hb_002174_040 |
0.137611203 |
- |
- |
DNAJ heat shock family protein [Theobroma cacao] |
| 4 |
Hb_121881_010 |
0.137839225 |
- |
- |
hypothetical protein POPTR_0015s15770g [Populus trichocarpa] |
| 5 |
Hb_008566_040 |
0.1435588104 |
- |
- |
PREDICTED: uncharacterized protein LOC105647590 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003935_060 |
0.147958562 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 7 |
Hb_001102_020 |
0.1483825125 |
- |
- |
solanesyl diphosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_004088_040 |
0.1517587629 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' iota isoform [Jatropha curcas] |
| 9 |
Hb_000300_200 |
0.1544989904 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_000244_180 |
0.1544997366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004223_050 |
0.1584673811 |
- |
- |
hexokinase [Manihot esculenta] |
| 12 |
Hb_000359_290 |
0.158605328 |
- |
- |
Triose phosphate/phosphate translocator, non-green plastid, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_001633_200 |
0.1590535654 |
- |
- |
hypothetical protein B456_005G137400 [Gossypium raimondii] |
| 14 |
Hb_000765_060 |
0.1607550394 |
- |
- |
Alcohol dehydrogenase-like 6 [Glycine soja] |
| 15 |
Hb_000300_600 |
0.1617640437 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 16 |
Hb_000022_200 |
0.162773545 |
- |
- |
PREDICTED: uncharacterized protein LOC105638292 [Jatropha curcas] |
| 17 |
Hb_000721_030 |
0.1637754526 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 18 |
Hb_000920_010 |
0.1675346209 |
- |
- |
hypothetical protein PRUPE_ppa013940mg [Prunus persica] |
| 19 |
Hb_000890_130 |
0.1693843243 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |
| 20 |
Hb_000108_050 |
0.1702619939 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |