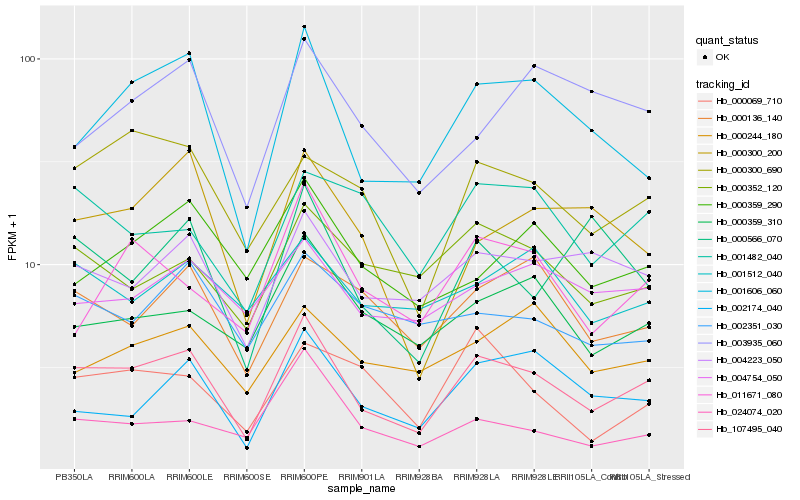
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_107495_040 |
0.0 |
- |
- |
PREDICTED: 12-oxophytodienoate reductase 2-like [Citrus sinensis] |
| 2 |
Hb_001606_060 |
0.128840726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004223_050 |
0.146032975 |
- |
- |
hexokinase [Manihot esculenta] |
| 4 |
Hb_000300_690 |
0.1549954049 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 5 |
Hb_002174_040 |
0.1554884364 |
- |
- |
DNAJ heat shock family protein [Theobroma cacao] |
| 6 |
Hb_000359_310 |
0.1570947986 |
- |
- |
hypothetical protein JCGZ_03934 [Jatropha curcas] |
| 7 |
Hb_000352_120 |
0.1597483366 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 8 |
Hb_000300_200 |
0.1600149913 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 9 |
Hb_000136_140 |
0.1609691401 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 10 |
Hb_011671_080 |
0.165874394 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein kinase TDR [Jatropha curcas] |
| 11 |
Hb_024074_020 |
0.166334314 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g09600 [Jatropha curcas] |
| 12 |
Hb_000359_290 |
0.1663556216 |
- |
- |
Triose phosphate/phosphate translocator, non-green plastid, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_004754_050 |
0.1675855567 |
- |
- |
PREDICTED: uncharacterized protein LOC104433925 [Eucalyptus grandis] |
| 14 |
Hb_002351_030 |
0.1697612577 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_000244_180 |
0.1698483422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001512_040 |
0.1699749983 |
- |
- |
PREDICTED: probable methyltransferase PMT9 [Jatropha curcas] |
| 17 |
Hb_003935_060 |
0.1712572915 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 18 |
Hb_000566_070 |
0.1718896706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001482_040 |
0.1723052513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000069_710 |
0.1748006608 |
- |
- |
PREDICTED: uncharacterized protein LOC105642890 [Jatropha curcas] |