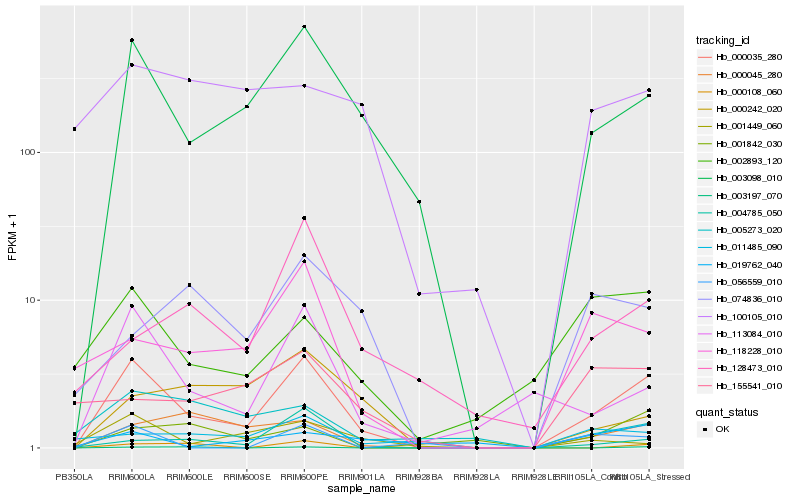
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_280 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 2 |
Hb_003098_010 |
0.1967548564 |
- |
- |
- |
| 3 |
Hb_113084_010 |
0.2747683546 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 4 |
Hb_118228_010 |
0.3150044263 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 5 |
Hb_000045_280 |
0.3163461079 |
- |
- |
hypothetical protein JCGZ_04076 [Jatropha curcas] |
| 6 |
Hb_002893_120 |
0.3238627361 |
- |
- |
PREDICTED: uncharacterized protein LOC105637692 [Jatropha curcas] |
| 7 |
Hb_004785_050 |
0.3248283365 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 8 |
Hb_001449_060 |
0.3249343744 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 9 |
Hb_000108_060 |
0.3358315614 |
- |
- |
hypothetical protein POPTR_0010s07820g [Populus trichocarpa] |
| 10 |
Hb_128473_010 |
0.3424300389 |
- |
- |
- |
| 11 |
Hb_056559_010 |
0.3448236914 |
- |
- |
PREDICTED: uncharacterized protein LOC104088595 [Nicotiana tomentosiformis] |
| 12 |
Hb_019762_040 |
0.3457567867 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001842_030 |
0.3489227609 |
- |
- |
PREDICTED: histone deacetylase 9 [Jatropha curcas] |
| 14 |
Hb_011485_090 |
0.3492242251 |
- |
- |
- |
| 15 |
Hb_000242_020 |
0.3500496841 |
- |
- |
hypothetical protein CICLE_v100246891mg, partial [Citrus clementina] |
| 16 |
Hb_074836_010 |
0.3508122907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003197_070 |
0.3549343699 |
- |
- |
PREDICTED: uncharacterized protein LOC104884990 [Beta vulgaris subsp. vulgaris] |
| 18 |
Hb_155541_010 |
0.3552874564 |
- |
- |
- |
| 19 |
Hb_100105_010 |
0.3561095654 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 20 |
Hb_005273_020 |
0.3562377498 |
- |
- |
- |