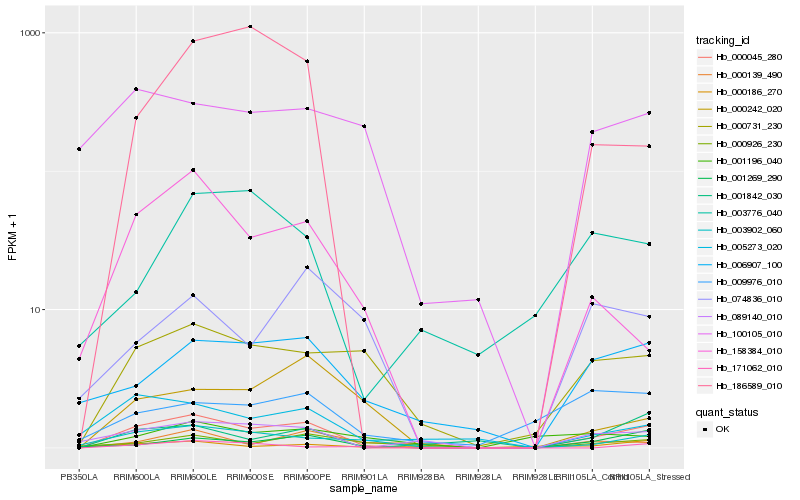
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000045_280 |
0.0 |
- |
- |
hypothetical protein JCGZ_04076 [Jatropha curcas] |
| 2 |
Hb_003902_060 |
0.2347911531 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 3 |
Hb_089140_010 |
0.2530973065 |
- |
- |
- |
| 4 |
Hb_000731_230 |
0.2558379153 |
- |
- |
PREDICTED: abnormal spindle-like microcephaly-associated protein homolog [Jatropha curcas] |
| 5 |
Hb_006907_100 |
0.2621214413 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 6 |
Hb_001269_290 |
0.2622436868 |
- |
- |
PREDICTED: uncharacterized protein LOC103438859 [Malus domestica] |
| 7 |
Hb_158384_010 |
0.2728464376 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 8 |
Hb_186589_010 |
0.2760752172 |
- |
- |
PREDICTED: developmentally-regulated G-protein 3 [Nelumbo nucifera] |
| 9 |
Hb_005273_020 |
0.2775272951 |
- |
- |
- |
| 10 |
Hb_001842_030 |
0.2843674693 |
- |
- |
PREDICTED: histone deacetylase 9 [Jatropha curcas] |
| 11 |
Hb_000926_230 |
0.2861756082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000242_020 |
0.2903095065 |
- |
- |
hypothetical protein CICLE_v100246891mg, partial [Citrus clementina] |
| 13 |
Hb_100105_010 |
0.2955740662 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 14 |
Hb_074836_010 |
0.3011059006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000139_490 |
0.3073797313 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 16 |
Hb_171062_010 |
0.307639982 |
- |
- |
hypothetical protein VITISV_036059 [Vitis vinifera] |
| 17 |
Hb_000186_270 |
0.3094555515 |
- |
- |
hypothetical protein JCGZ_02112 [Jatropha curcas] |
| 18 |
Hb_009976_010 |
0.309725066 |
- |
- |
PREDICTED: uncharacterized protein LOC105630216 [Jatropha curcas] |
| 19 |
Hb_003776_040 |
0.312555241 |
- |
- |
PREDICTED: RING finger protein 165-like isoform X2 [Populus euphratica] |
| 20 |
Hb_001196_040 |
0.3128691726 |
- |
- |
PREDICTED: uncharacterized protein LOC104100193 [Nicotiana tomentosiformis] |