| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
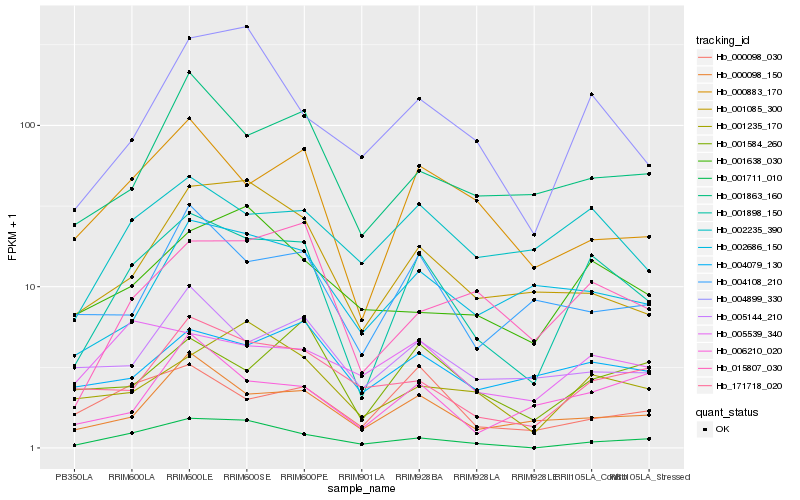
Hb_001898_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 2 |
Hb_000883_170 |
0.1838984709 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 3 |
Hb_000098_150 |
0.1841202836 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 4 |
Hb_002235_390 |
0.1877923884 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 5 |
Hb_015807_030 |
0.1935837781 |
- |
- |
PREDICTED: protein NDR1-like [Jatropha curcas] |
| 6 |
Hb_001711_010 |
0.1945222944 |
- |
- |
- |
| 7 |
Hb_001863_160 |
0.1964376459 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 8 |
Hb_004899_330 |
0.2005576635 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 9 |
Hb_000098_030 |
0.2012724556 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |
| 10 |
Hb_001085_300 |
0.2047762247 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 11 |
Hb_001235_170 |
0.2055778452 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 12 |
Hb_001584_260 |
0.2069222164 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_171718_020 |
0.2069727484 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 14 |
Hb_004108_210 |
0.2074247035 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 15 |
Hb_002686_150 |
0.2081318327 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 16 |
Hb_005144_210 |
0.2094234114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_004079_130 |
0.2095756996 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 18 |
Hb_005539_340 |
0.2154476206 |
- |
- |
PREDICTED: CDT1-like protein a, chloroplastic [Jatropha curcas] |
| 19 |
Hb_006210_020 |
0.2163782808 |
- |
- |
PREDICTED: calcium sensing receptor, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001638_030 |
0.2170302933 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |