| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
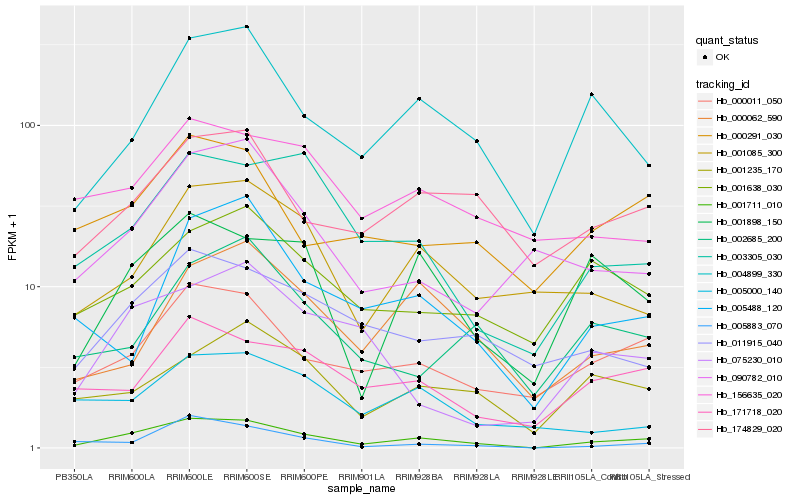
Hb_001711_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000011_050 |
0.1775246709 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 3 |
Hb_004899_330 |
0.1880629056 |
transcription factor |
TF Family: Tify |
plastid jasmonates ZIM-domain protein [Hevea brasiliensis] |
| 4 |
Hb_001898_150 |
0.1945222944 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 5 |
Hb_011915_040 |
0.1982847702 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 6 |
Hb_171718_020 |
0.1994368019 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 7 |
Hb_003305_030 |
0.1994377314 |
- |
- |
- |
| 8 |
Hb_001085_300 |
0.2015718826 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 5 [Jatropha curcas] |
| 9 |
Hb_174829_020 |
0.2028826998 |
transcription factor |
TF Family: C2C2-Dof |
cyclic dof factor 2 [Jatropha curcas] |
| 10 |
Hb_000291_030 |
0.2063063094 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1-like [Eucalyptus grandis] |
| 11 |
Hb_005883_070 |
0.208191585 |
- |
- |
PREDICTED: probable myosin-binding protein 5 [Jatropha curcas] |
| 12 |
Hb_001235_170 |
0.2093270242 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 13 |
Hb_000062_590 |
0.2099860529 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 14 |
Hb_001638_030 |
0.2118274896 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 15 |
Hb_005488_120 |
0.2136147794 |
- |
- |
PREDICTED: DNA polymerase delta subunit 4 [Jatropha curcas] |
| 16 |
Hb_090782_010 |
0.213751006 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 17 |
Hb_002685_200 |
0.2170322069 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 18 |
Hb_075230_010 |
0.2174764614 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like [Glycine max] |
| 19 |
Hb_156635_020 |
0.217931214 |
- |
- |
PREDICTED: L-lactate dehydrogenase B [Jatropha curcas] |
| 20 |
Hb_005000_140 |
0.2185643537 |
- |
- |
PREDICTED: uncharacterized protein LOC105637876 isoform X1 [Jatropha curcas] |