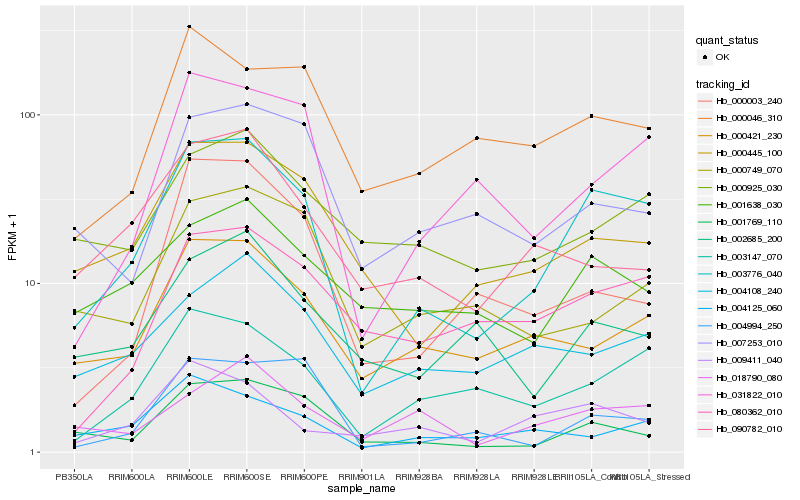
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003776_040 |
0.0 |
- |
- |
PREDICTED: RING finger protein 165-like isoform X2 [Populus euphratica] |
| 2 |
Hb_003147_070 |
0.16888154 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like [Jatropha curcas] |
| 3 |
Hb_031822_010 |
0.1808716314 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 4 |
Hb_000445_100 |
0.1854459376 |
- |
- |
PREDICTED: glutathione S-transferase TCHQD [Jatropha curcas] |
| 5 |
Hb_001769_110 |
0.1883075648 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |
| 6 |
Hb_000421_230 |
0.1890999685 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 7 |
Hb_080362_010 |
0.2012873925 |
- |
- |
hypothetical protein JCGZ_14167 [Jatropha curcas] |
| 8 |
Hb_004125_060 |
0.2046088623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004108_240 |
0.204662078 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 10 |
Hb_018790_080 |
0.2083484536 |
- |
- |
hypothetical protein POPTR_0011s06335g [Populus trichocarpa] |
| 11 |
Hb_004994_250 |
0.2087412396 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB6 [Jatropha curcas] |
| 12 |
Hb_000046_310 |
0.2092315092 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 13 |
Hb_001638_030 |
0.2095940792 |
desease resistance |
Gene Name: RPW8 |
PREDICTED: probable disease resistance protein At4g33300 [Jatropha curcas] |
| 14 |
Hb_009411_040 |
0.2127256994 |
- |
- |
- |
| 15 |
Hb_000925_030 |
0.2145543414 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_002685_200 |
0.2155543758 |
- |
- |
PREDICTED: uncharacterized protein LOC105632583 [Jatropha curcas] |
| 17 |
Hb_000003_240 |
0.2163199841 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007253_010 |
0.2177160341 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 19 |
Hb_090782_010 |
0.2190926 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 20 |
Hb_000749_070 |
0.2207588357 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |