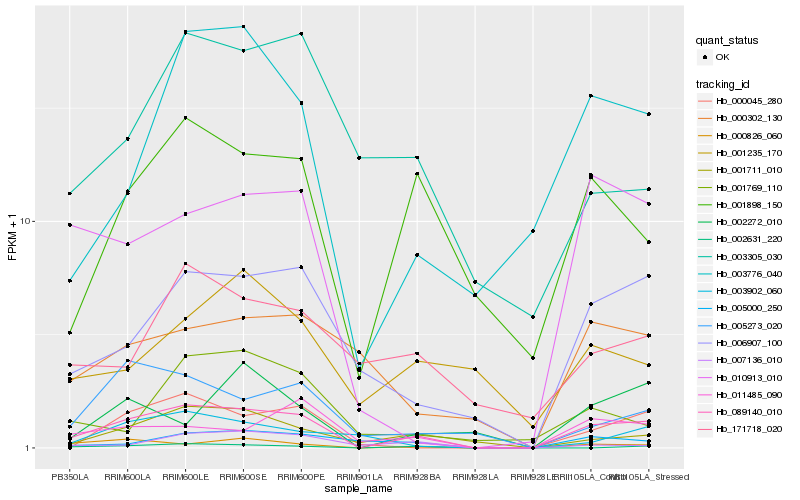
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089140_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_006907_100 |
0.2168635501 |
- |
- |
PREDICTED: tropinone reductase-like 3 [Jatropha curcas] |
| 3 |
Hb_001898_150 |
0.2280480693 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 4 |
Hb_000826_060 |
0.2336502151 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 5 |
Hb_001711_010 |
0.2365989762 |
- |
- |
- |
| 6 |
Hb_003776_040 |
0.2373797322 |
- |
- |
PREDICTED: RING finger protein 165-like isoform X2 [Populus euphratica] |
| 7 |
Hb_010913_010 |
0.2484219885 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 8 |
Hb_005273_020 |
0.2496746891 |
- |
- |
- |
| 9 |
Hb_011485_090 |
0.2504590506 |
- |
- |
- |
| 10 |
Hb_000045_280 |
0.2530973065 |
- |
- |
hypothetical protein JCGZ_04076 [Jatropha curcas] |
| 11 |
Hb_005000_250 |
0.2548620942 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 4, mitochondrial [Vitis vinifera] |
| 12 |
Hb_002631_220 |
0.2552455998 |
- |
- |
Patellin-4, putative [Ricinus communis] |
| 13 |
Hb_171718_020 |
0.2636713079 |
- |
- |
hypothetical protein JCGZ_00274 [Jatropha curcas] |
| 14 |
Hb_000302_130 |
0.268066315 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 1 [Jatropha curcas] |
| 15 |
Hb_003902_060 |
0.2728325917 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 16 |
Hb_002272_010 |
0.2761242507 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003305_030 |
0.2774353758 |
- |
- |
- |
| 18 |
Hb_007136_010 |
0.2780063316 |
desease resistance |
Gene Name: UBA |
PREDICTED: ATPase WRNIP1 [Jatropha curcas] |
| 19 |
Hb_001235_170 |
0.280892541 |
- |
- |
PREDICTED: putative glucuronosyltransferase PGSIP8 [Jatropha curcas] |
| 20 |
Hb_001769_110 |
0.2824932433 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL7 [Jatropha curcas] |