| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
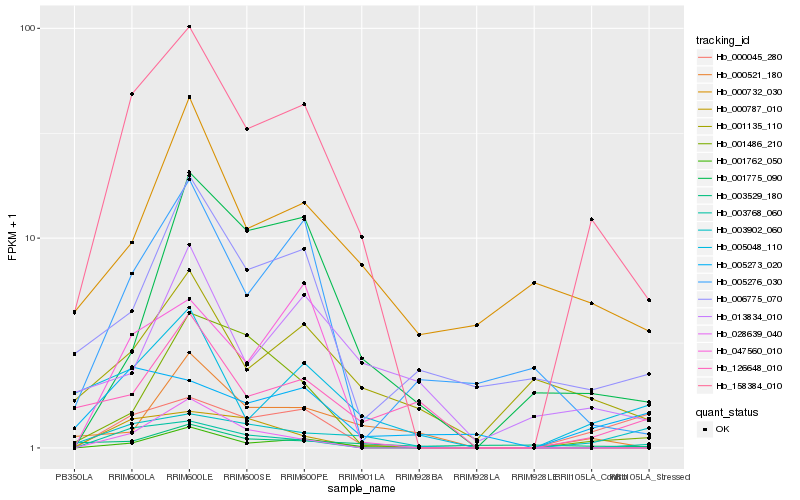
Hb_158384_010 |
0.0 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 2 |
Hb_001762_050 |
0.231654406 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 3 |
Hb_005048_110 |
0.2512456686 |
- |
- |
- |
| 4 |
Hb_003768_060 |
0.2614822061 |
- |
- |
PREDICTED: transcription initiation factor IIB-like [Jatropha curcas] |
| 5 |
Hb_001135_110 |
0.2669727865 |
- |
- |
- |
| 6 |
Hb_006775_070 |
0.2716377266 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_003902_060 |
0.2718869958 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 8 |
Hb_000045_280 |
0.2728464376 |
- |
- |
hypothetical protein JCGZ_04076 [Jatropha curcas] |
| 9 |
Hb_000521_180 |
0.2749254337 |
- |
- |
PREDICTED: uncharacterized protein LOC105650335 [Jatropha curcas] |
| 10 |
Hb_000732_030 |
0.2754844982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001775_090 |
0.2778809815 |
- |
- |
hypothetical protein JCGZ_11262 [Jatropha curcas] |
| 12 |
Hb_001486_210 |
0.2788044889 |
- |
- |
- |
| 13 |
Hb_126648_010 |
0.2835090605 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 33 homolog [Jatropha curcas] |
| 14 |
Hb_005276_030 |
0.2847836414 |
- |
- |
PREDICTED: probable inactive patatin-like protein 9 [Jatropha curcas] |
| 15 |
Hb_013834_010 |
0.2856553223 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Jatropha curcas] |
| 16 |
Hb_000787_010 |
0.2858961823 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 17 |
Hb_047560_010 |
0.2871592527 |
- |
- |
PREDICTED: GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 18 |
Hb_005273_020 |
0.2896100848 |
- |
- |
- |
| 19 |
Hb_028639_040 |
0.2945895373 |
- |
- |
PREDICTED: uncharacterized protein LOC105761689 [Gossypium raimondii] |
| 20 |
Hb_003529_180 |
0.2977001802 |
- |
- |
hypothetical protein JCGZ_00222 [Jatropha curcas] |