| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
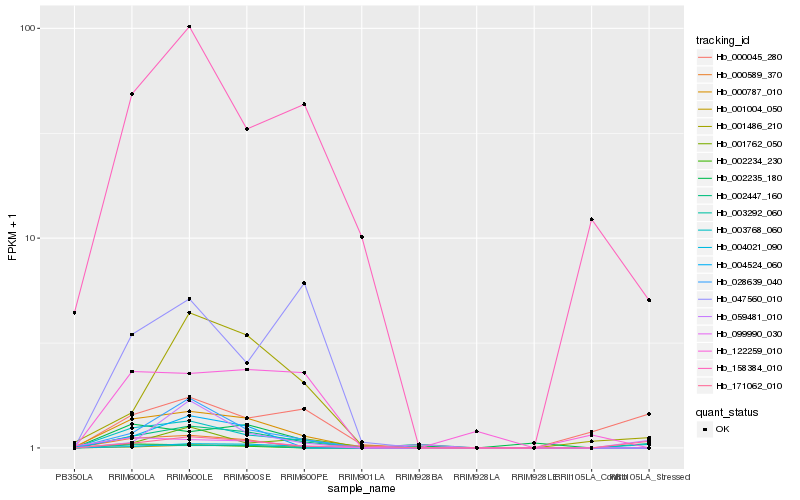
Hb_003768_060 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor IIB-like [Jatropha curcas] |
| 2 |
Hb_000787_010 |
0.1910981813 |
- |
- |
PREDICTED: tryptophan synthase beta chain 1 [Jatropha curcas] |
| 3 |
Hb_002447_160 |
0.2047405367 |
- |
- |
- |
| 4 |
Hb_028639_040 |
0.2341585978 |
- |
- |
PREDICTED: uncharacterized protein LOC105761689 [Gossypium raimondii] |
| 5 |
Hb_158384_010 |
0.2614822061 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 6 |
Hb_004021_090 |
0.2637156553 |
- |
- |
hypothetical protein JCGZ_09233 [Jatropha curcas] |
| 7 |
Hb_099990_030 |
0.2701592028 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 8 |
Hb_000589_370 |
0.2854326442 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 33 [Jatropha curcas] |
| 9 |
Hb_001486_210 |
0.3000119621 |
- |
- |
- |
| 10 |
Hb_002234_230 |
0.3000373627 |
- |
- |
hypothetical protein JCGZ_26141 [Jatropha curcas] |
| 11 |
Hb_001762_050 |
0.3049321841 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 12 |
Hb_122259_010 |
0.3093916336 |
- |
- |
- |
| 13 |
Hb_171062_010 |
0.3112302478 |
- |
- |
hypothetical protein VITISV_036059 [Vitis vinifera] |
| 14 |
Hb_003292_060 |
0.3137619775 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38) [Jatropha curcas] |
| 15 |
Hb_002235_180 |
0.3153408007 |
transcription factor |
TF Family: ERF |
PREDICTED: uncharacterized protein LOC105133481 [Populus euphratica] |
| 16 |
Hb_000045_280 |
0.315663363 |
- |
- |
hypothetical protein JCGZ_04076 [Jatropha curcas] |
| 17 |
Hb_047560_010 |
0.3223456808 |
- |
- |
PREDICTED: GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase [Jatropha curcas] |
| 18 |
Hb_059481_010 |
0.3235018845 |
- |
- |
hypothetical protein POPTR_0008s06860g [Populus trichocarpa] |
| 19 |
Hb_001004_050 |
0.3276563001 |
- |
- |
PREDICTED: uncharacterized protein LOC103335231 [Prunus mume] |
| 20 |
Hb_004524_060 |
0.3282938299 |
- |
- |
hypothetical protein VITISV_039334 [Vitis vinifera] |