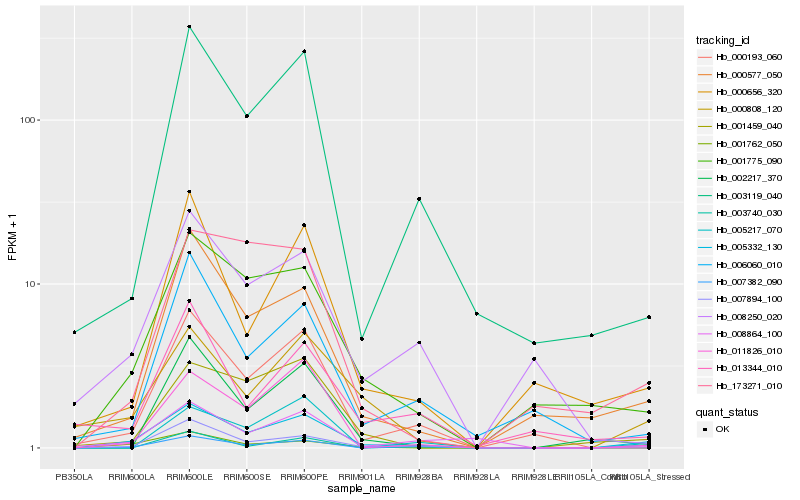
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005332_130 |
0.0 |
- |
- |
Mavicyanin, putative [Ricinus communis] |
| 2 |
Hb_007382_090 |
0.1631677768 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_000808_120 |
0.1884456732 |
- |
- |
- |
| 4 |
Hb_001775_090 |
0.1918025371 |
- |
- |
hypothetical protein JCGZ_11262 [Jatropha curcas] |
| 5 |
Hb_000577_050 |
0.1969931601 |
- |
- |
- |
| 6 |
Hb_007894_100 |
0.2033023219 |
- |
- |
PREDICTED: uncharacterized protein LOC105632952 [Jatropha curcas] |
| 7 |
Hb_003119_040 |
0.2055580424 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 8 |
Hb_000193_060 |
0.2074548836 |
- |
- |
PREDICTED: F-box protein At4g35930 [Jatropha curcas] |
| 9 |
Hb_000656_320 |
0.2123095092 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 10 |
Hb_011826_010 |
0.2201026649 |
- |
- |
Disease resistance protein RPM1, putative [Ricinus communis] |
| 11 |
Hb_001459_040 |
0.221666633 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 12 |
Hb_006060_010 |
0.2250954687 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 13 |
Hb_001762_050 |
0.2254841754 |
- |
- |
PREDICTED: xylem cysteine proteinase 1 [Jatropha curcas] |
| 14 |
Hb_002217_370 |
0.2341342545 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105643685 [Jatropha curcas] |
| 15 |
Hb_008864_100 |
0.2364662526 |
- |
- |
PREDICTED: uncharacterized protein LOC105631832 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005217_070 |
0.2371509949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003740_030 |
0.2398239522 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 18 |
Hb_008250_020 |
0.240892743 |
- |
- |
PREDICTED: U-box domain-containing protein 15-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_013344_010 |
0.2420489885 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 [Jatropha curcas] |
| 20 |
Hb_173271_010 |
0.243452582 |
- |
- |
Disease resistance protein RPP13, putative [Ricinus communis] |