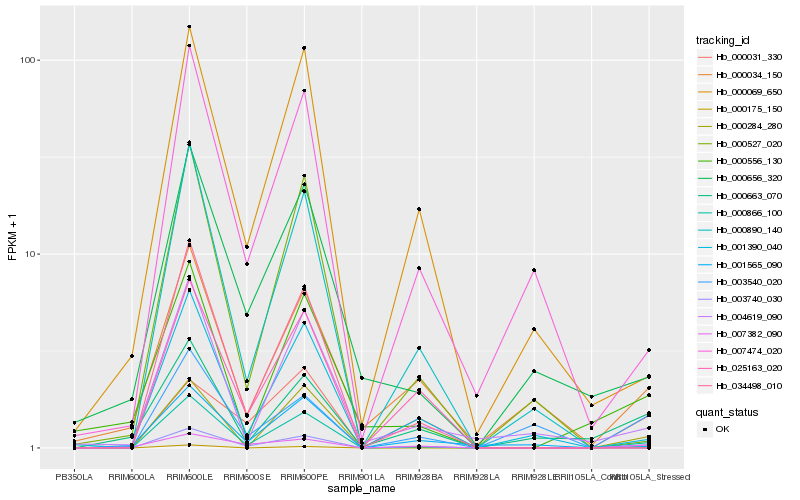
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003740_030 |
0.0 |
transcription factor |
TF Family: RAV |
DNA-binding protein RAV1, putative [Ricinus communis] |
| 2 |
Hb_000866_100 |
0.1129535884 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_007382_090 |
0.1632910564 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_001390_040 |
0.1903434999 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 5 |
Hb_007474_020 |
0.2073552087 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 6 |
Hb_034498_010 |
0.2107656997 |
- |
- |
PREDICTED: uncharacterized protein LOC105636842 [Jatropha curcas] |
| 7 |
Hb_000034_150 |
0.2120003281 |
- |
- |
PREDICTED: kinesin-like protein BC2 [Jatropha curcas] |
| 8 |
Hb_000069_650 |
0.2135168631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000284_280 |
0.2158692892 |
- |
- |
- |
| 10 |
Hb_000031_330 |
0.2170147528 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0157s00200g, partial [Populus trichocarpa] |
| 11 |
Hb_004619_090 |
0.2187041975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000663_070 |
0.2200165826 |
- |
- |
cyclin A, putative [Ricinus communis] |
| 13 |
Hb_003540_020 |
0.2202024101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_025163_020 |
0.2202549214 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 15 |
Hb_000890_140 |
0.223343446 |
- |
- |
PREDICTED: uncharacterized protein LOC105646185 [Jatropha curcas] |
| 16 |
Hb_001565_090 |
0.2250341115 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000175_150 |
0.2267327571 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein B456_009G311700 [Gossypium raimondii] |
| 18 |
Hb_000527_020 |
0.2268667011 |
- |
- |
PREDICTED: uncharacterized protein LOC105649490 [Jatropha curcas] |
| 19 |
Hb_000556_130 |
0.2277684645 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 20 |
Hb_000656_320 |
0.2281376121 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |