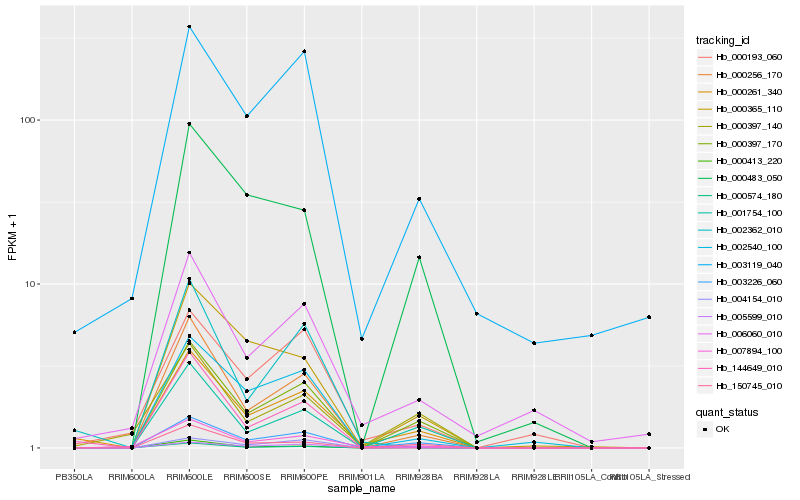
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_340 |
0.0 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 2 |
Hb_000397_170 |
0.1415614565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003226_060 |
0.1424333134 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 4 |
Hb_000574_180 |
0.1546573871 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 5 |
Hb_000256_170 |
0.1594627105 |
- |
- |
PREDICTED: uncharacterized protein LOC105638320 [Jatropha curcas] |
| 6 |
Hb_005599_010 |
0.1614822332 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 7 |
Hb_007894_100 |
0.1685408832 |
- |
- |
PREDICTED: uncharacterized protein LOC105632952 [Jatropha curcas] |
| 8 |
Hb_001754_100 |
0.1716874308 |
- |
- |
PREDICTED: peroxidase 5-like [Jatropha curcas] |
| 9 |
Hb_000413_220 |
0.1740162185 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 10 |
Hb_150745_010 |
0.1782537147 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 11 |
Hb_003119_040 |
0.1802917225 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 12 |
Hb_004154_010 |
0.1816963666 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 13 |
Hb_002362_010 |
0.1888461143 |
- |
- |
hypothetical protein JCGZ_12328 [Jatropha curcas] |
| 14 |
Hb_006060_010 |
0.1890434592 |
- |
- |
PREDICTED: cyclin-A2-2-like [Jatropha curcas] |
| 15 |
Hb_002540_100 |
0.1892603736 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 16 |
Hb_144649_010 |
0.1896835002 |
- |
- |
60S ribosomal protein L34A [Hevea brasiliensis] |
| 17 |
Hb_000193_060 |
0.1912787696 |
- |
- |
PREDICTED: F-box protein At4g35930 [Jatropha curcas] |
| 18 |
Hb_000397_140 |
0.1919555444 |
transcription factor |
TF Family: PLATZ |
hypothetical protein JCGZ_16980 [Jatropha curcas] |
| 19 |
Hb_000483_050 |
0.1920626061 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 20 |
Hb_000365_110 |
0.1931077708 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |