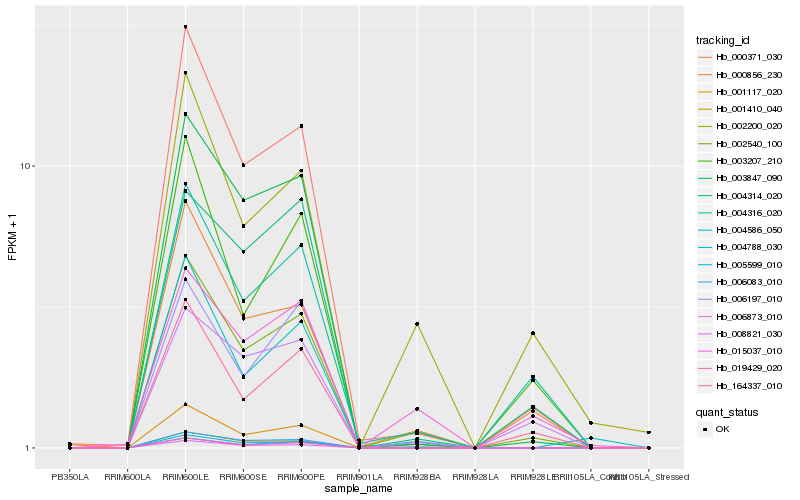
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002540_100 |
0.0 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 2 |
Hb_000371_030 |
0.09112003 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: anthocyanidin 3-O-glucosyltransferase 5 [Prunus mume] |
| 3 |
Hb_000856_230 |
0.1038581747 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 4 |
Hb_004316_020 |
0.1133773663 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1-like [Jatropha curcas] |
| 5 |
Hb_003847_090 |
0.1164720499 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 6 |
Hb_005599_010 |
0.1171192508 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 7 |
Hb_006873_010 |
0.1251167248 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_019429_020 |
0.1256561011 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_006197_010 |
0.1279092218 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 10 |
Hb_001117_020 |
0.1286942333 |
- |
- |
hypothetical protein CICLE_v10008579mg [Citrus clementina] |
| 11 |
Hb_164337_010 |
0.1313143692 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 [Jatropha curcas] |
| 12 |
Hb_015037_010 |
0.1317433499 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 13 |
Hb_004788_030 |
0.1317561031 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_008821_030 |
0.1321578192 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 15 |
Hb_004586_050 |
0.1332803482 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 16 |
Hb_002200_020 |
0.1343795595 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein RCOM_0137680 [Ricinus communis] |
| 17 |
Hb_004314_020 |
0.1361302876 |
- |
- |
Auxin-responsive GH3 family protein [Theobroma cacao] |
| 18 |
Hb_006083_010 |
0.1382745635 |
transcription factor |
TF Family: B3 |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_003207_210 |
0.1401115567 |
- |
- |
PREDICTED: uncharacterized protein LOC105645885 [Jatropha curcas] |
| 20 |
Hb_001410_040 |
0.1403431757 |
- |
- |
Protein ABIL2, putative [Ricinus communis] |