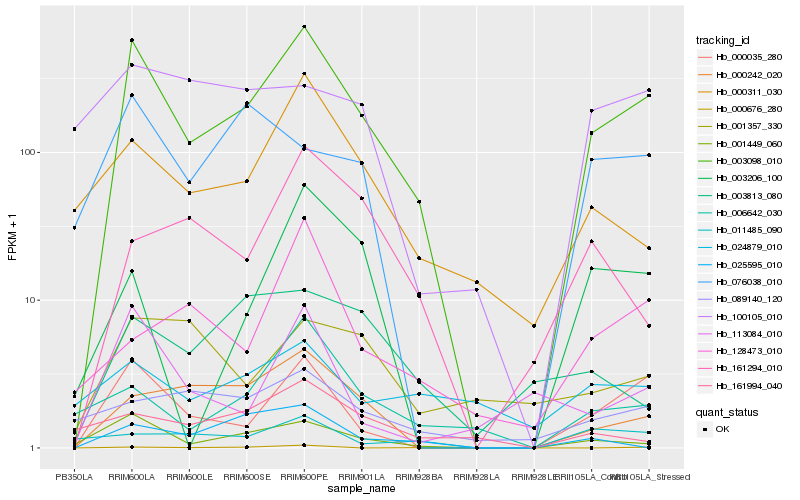
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003098_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000035_280 |
0.1967548564 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 3 |
Hb_001449_060 |
0.2044961247 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 4 |
Hb_000242_020 |
0.258946096 |
- |
- |
hypothetical protein CICLE_v100246891mg, partial [Citrus clementina] |
| 5 |
Hb_000311_030 |
0.2719217909 |
- |
- |
hypothetical protein B456_002G227600 [Gossypium raimondii] |
| 6 |
Hb_006642_030 |
0.2790428526 |
- |
- |
PREDICTED: germin-like protein subfamily 3 member 2 [Jatropha curcas] |
| 7 |
Hb_113084_010 |
0.2876098937 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 8 |
Hb_089140_120 |
0.2962895397 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 9 |
Hb_076038_010 |
0.3011623615 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_011485_090 |
0.3049508215 |
- |
- |
- |
| 11 |
Hb_003206_100 |
0.3053359444 |
- |
- |
- |
| 12 |
Hb_161294_010 |
0.3059375675 |
- |
- |
PREDICTED: serine/threonine-protein kinase ppk15 isoform X2 [Jatropha curcas] |
| 13 |
Hb_128473_010 |
0.3063253907 |
- |
- |
- |
| 14 |
Hb_024879_010 |
0.3075839581 |
- |
- |
unknown [Glycine max] |
| 15 |
Hb_100105_010 |
0.3103679599 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 16 |
Hb_161994_040 |
0.3112078064 |
- |
- |
- |
| 17 |
Hb_003813_080 |
0.3122207161 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 18 |
Hb_025595_010 |
0.3123661853 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 19 |
Hb_001357_330 |
0.3146039735 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 20 |
Hb_000676_280 |
0.3188158947 |
- |
- |
ATP binding protein, putative [Ricinus communis] |