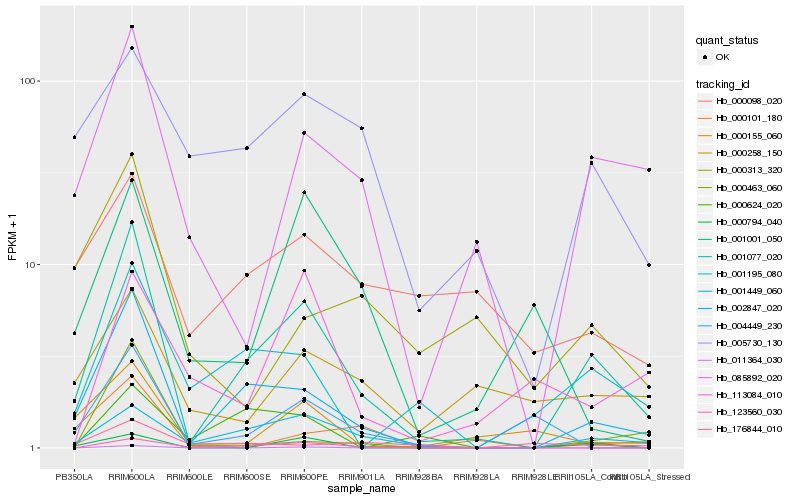
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001077_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_23832 [Jatropha curcas] |
| 2 |
Hb_002847_020 |
0.1546673784 |
- |
- |
hypothetical protein JCGZ_23832 [Jatropha curcas] |
| 3 |
Hb_001449_060 |
0.2348413053 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 4 |
Hb_085892_020 |
0.2720839585 |
- |
- |
- |
| 5 |
Hb_001195_080 |
0.2953065446 |
- |
- |
PREDICTED: uncharacterized protein LOC105633814 [Jatropha curcas] |
| 6 |
Hb_123560_030 |
0.3097905144 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000794_040 |
0.3131028382 |
- |
- |
PREDICTED: RING-H2 finger protein ATL29-like [Jatropha curcas] |
| 8 |
Hb_005730_130 |
0.3183032181 |
- |
- |
PRA1 family protein [Theobroma cacao] |
| 9 |
Hb_000155_060 |
0.3320375016 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_004449_230 |
0.337285541 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_000258_150 |
0.3389458229 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 12 |
Hb_000101_180 |
0.3407817674 |
- |
- |
- |
| 13 |
Hb_000098_020 |
0.3530372708 |
transcription factor |
TF Family: GRAS |
GRAS family transcription factor family protein [Theobroma cacao] |
| 14 |
Hb_000624_020 |
0.3539811394 |
transcription factor |
TF Family: NAC |
NAC transcription factor 004 [Jatropha curcas] |
| 15 |
Hb_000313_320 |
0.354737307 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 16 |
Hb_001001_050 |
0.357299353 |
- |
- |
CXE protein [Hevea brasiliensis] |
| 17 |
Hb_176844_010 |
0.358820094 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000463_060 |
0.3628545518 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 19 |
Hb_011364_030 |
0.3643528804 |
- |
- |
PREDICTED: U-box domain-containing protein 51 [Jatropha curcas] |
| 20 |
Hb_113084_010 |
0.3650413061 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |