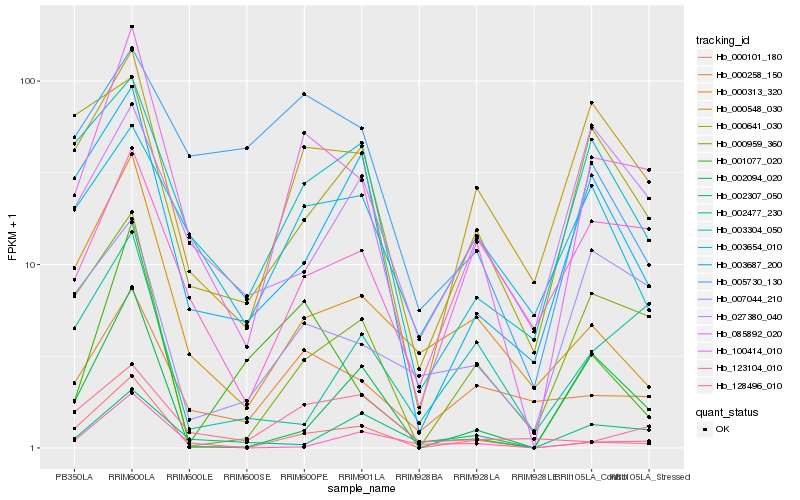
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_085892_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000548_030 |
0.197655793 |
rubber biosynthesis |
Gene Name: CPT5 |
PREDICTED: dehydrodolichyl diphosphate synthase 6-like [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_000258_150 |
0.2219727112 |
transcription factor |
TF Family: zf-HD |
PREDICTED: zinc-finger homeodomain protein 11-like [Jatropha curcas] |
| 4 |
Hb_000959_360 |
0.2278649641 |
- |
- |
transporter, putative [Ricinus communis] |
| 5 |
Hb_000101_180 |
0.2309818381 |
- |
- |
- |
| 6 |
Hb_003304_050 |
0.2371210086 |
- |
- |
PREDICTED: uncharacterized protein LOC105633679 isoform X1 [Jatropha curcas] |
| 7 |
Hb_100414_010 |
0.2393300443 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl pyrophosphate synthase [Hevea brasiliensis] |
| 8 |
Hb_000313_320 |
0.251494888 |
- |
- |
PREDICTED: protein kinase PINOID [Jatropha curcas] |
| 9 |
Hb_003687_200 |
0.2576185176 |
- |
- |
PREDICTED: uncharacterized protein LOC105632139 [Jatropha curcas] |
| 10 |
Hb_007044_210 |
0.2595404022 |
transcription factor |
TF Family: MYB |
PREDICTED: protein rough sheath 2 homolog [Populus euphratica] |
| 11 |
Hb_003654_010 |
0.2688626341 |
- |
- |
PREDICTED: probable polygalacturonase non-catalytic subunit JP650 [Jatropha curcas] |
| 12 |
Hb_002094_020 |
0.2693261303 |
- |
- |
PREDICTED: alpha-copaene synthase-like [Jatropha curcas] |
| 13 |
Hb_001077_020 |
0.2720839585 |
- |
- |
hypothetical protein JCGZ_23832 [Jatropha curcas] |
| 14 |
Hb_128496_010 |
0.2775940889 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_123104_010 |
0.281777948 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 16 |
Hb_000641_030 |
0.2824113282 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 17 |
Hb_005730_130 |
0.2875958082 |
- |
- |
PRA1 family protein [Theobroma cacao] |
| 18 |
Hb_027380_040 |
0.28913643 |
- |
- |
hypothetical protein B456_011G046500, partial [Gossypium raimondii] |
| 19 |
Hb_002307_050 |
0.2894341144 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002477_230 |
0.2898544704 |
- |
- |
PREDICTED: verprolin [Jatropha curcas] |