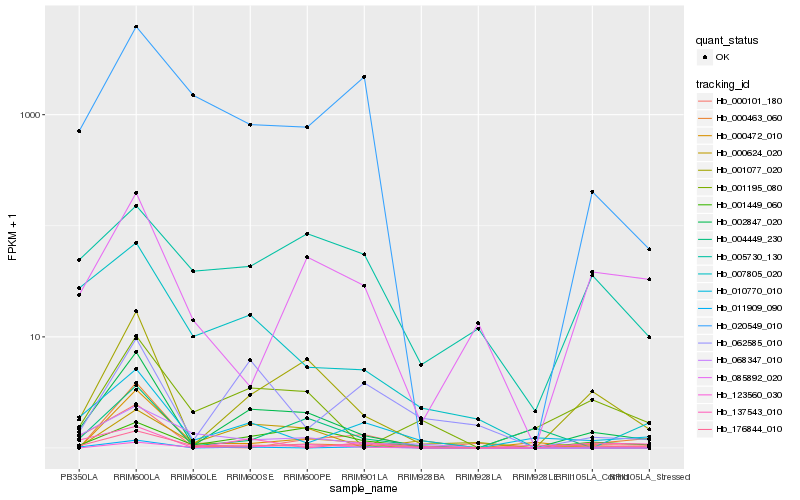
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002847_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_23832 [Jatropha curcas] |
| 2 |
Hb_001077_020 |
0.1546673784 |
- |
- |
hypothetical protein JCGZ_23832 [Jatropha curcas] |
| 3 |
Hb_001449_060 |
0.2790815994 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 4 |
Hb_001195_080 |
0.2822836239 |
- |
- |
PREDICTED: uncharacterized protein LOC105633814 [Jatropha curcas] |
| 5 |
Hb_010770_010 |
0.2937569344 |
- |
- |
- |
| 6 |
Hb_176844_010 |
0.3172013911 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000624_020 |
0.3251970993 |
transcription factor |
TF Family: NAC |
NAC transcription factor 004 [Jatropha curcas] |
| 8 |
Hb_007805_020 |
0.325445682 |
- |
- |
- |
| 9 |
Hb_085892_020 |
0.3273026818 |
- |
- |
- |
| 10 |
Hb_123560_030 |
0.3284357272 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000463_060 |
0.3312723258 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 12 |
Hb_068347_010 |
0.3333868834 |
- |
- |
- |
| 13 |
Hb_011909_090 |
0.3340488474 |
transcription factor |
TF Family: NOZZLE |
PREDICTED: uncharacterized protein LOC105633212 [Jatropha curcas] |
| 14 |
Hb_004449_230 |
0.3346559756 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_000472_010 |
0.3353270735 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 16 |
Hb_020549_010 |
0.3373780325 |
- |
- |
PREDICTED: uncharacterized protein LOC105638664 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000101_180 |
0.3451222511 |
- |
- |
- |
| 18 |
Hb_137543_010 |
0.3452601661 |
- |
- |
- |
| 19 |
Hb_005730_130 |
0.345745583 |
- |
- |
PRA1 family protein [Theobroma cacao] |
| 20 |
Hb_062585_010 |
0.3516145714 |
- |
- |
hypothetical protein JCGZ_19532 [Jatropha curcas] |