| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
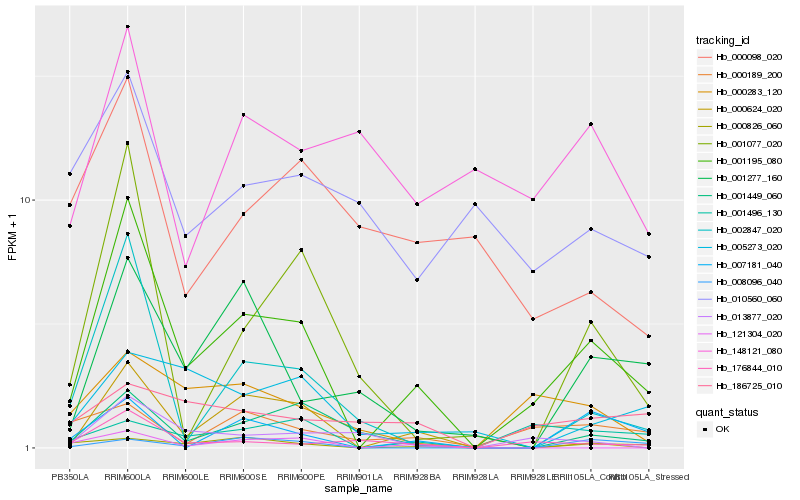
Hb_001195_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633814 [Jatropha curcas] |
| 2 |
Hb_000624_020 |
0.2747470195 |
transcription factor |
TF Family: NAC |
NAC transcription factor 004 [Jatropha curcas] |
| 3 |
Hb_176844_010 |
0.2811116038 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_002847_020 |
0.2822836239 |
- |
- |
hypothetical protein JCGZ_23832 [Jatropha curcas] |
| 5 |
Hb_001077_020 |
0.2953065446 |
- |
- |
hypothetical protein JCGZ_23832 [Jatropha curcas] |
| 6 |
Hb_001449_060 |
0.3004562712 |
- |
- |
kelch repeat protein, putative [Ricinus communis] |
| 7 |
Hb_007181_040 |
0.3006470943 |
- |
- |
- |
| 8 |
Hb_000826_060 |
0.3042793246 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 9 |
Hb_000283_120 |
0.3124823981 |
- |
- |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 10 |
Hb_001277_160 |
0.3144417089 |
- |
- |
hypothetical protein MIMGU_mgv1a019742mg, partial [Erythranthe guttata] |
| 11 |
Hb_013877_020 |
0.3191740612 |
- |
- |
Cytochrome P450 [Theobroma cacao] |
| 12 |
Hb_001496_130 |
0.3224619118 |
- |
- |
- |
| 13 |
Hb_000189_200 |
0.329359663 |
- |
- |
hypothetical protein JCGZ_02242 [Jatropha curcas] |
| 14 |
Hb_000098_020 |
0.3363782979 |
transcription factor |
TF Family: GRAS |
GRAS family transcription factor family protein [Theobroma cacao] |
| 15 |
Hb_121304_020 |
0.3366160347 |
- |
- |
- |
| 16 |
Hb_005273_020 |
0.3369088663 |
- |
- |
- |
| 17 |
Hb_186725_010 |
0.3378275041 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_148121_080 |
0.3428136472 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SHOOT GRAVITROPISM 5 [Populus euphratica] |
| 19 |
Hb_008096_040 |
0.3444979205 |
- |
- |
hypothetical protein PRUPE_ppa001351mg [Prunus persica] |
| 20 |
Hb_010560_060 |
0.3451234893 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |