| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
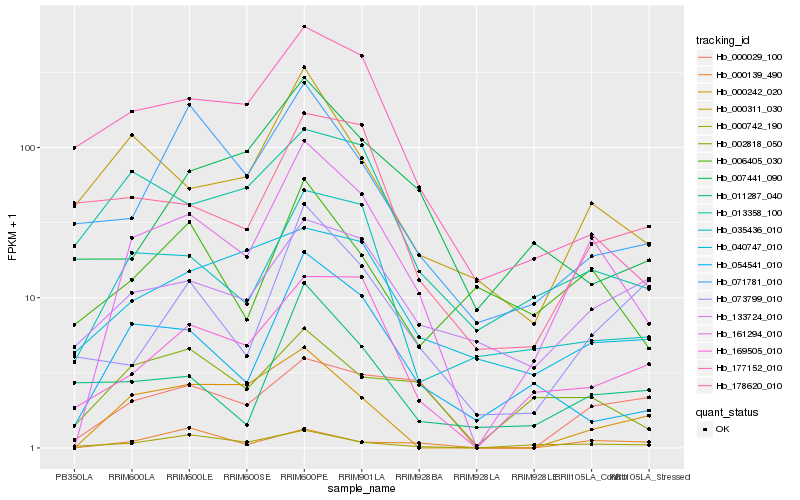
Hb_161294_010 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase ppk15 isoform X2 [Jatropha curcas] |
| 2 |
Hb_054541_010 |
0.2154368581 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X3 [Jatropha curcas] |
| 3 |
Hb_035436_010 |
0.2217098052 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 4 |
Hb_169505_010 |
0.2262840519 |
- |
- |
hypothetical protein JCGZ_04480 [Jatropha curcas] |
| 5 |
Hb_000742_190 |
0.2351066878 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000311_030 |
0.2393930045 |
- |
- |
hypothetical protein B456_002G227600 [Gossypium raimondii] |
| 7 |
Hb_002818_050 |
0.2429936927 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 8 |
Hb_013358_100 |
0.2479457718 |
- |
- |
unknown [Medicago truncatula] |
| 9 |
Hb_177152_010 |
0.2488496734 |
- |
- |
glycosyl hydrolase family 38 family protein [Populus trichocarpa] |
| 10 |
Hb_000242_020 |
0.252484317 |
- |
- |
hypothetical protein CICLE_v100246891mg, partial [Citrus clementina] |
| 11 |
Hb_133724_010 |
0.2606239594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_071781_010 |
0.2615546071 |
- |
- |
hypothetical protein POPTR_0010s13190g [Populus trichocarpa] |
| 13 |
Hb_006405_030 |
0.2622958349 |
- |
- |
hypothetical protein VITISV_043238 [Vitis vinifera] |
| 14 |
Hb_000139_490 |
0.2627914109 |
- |
- |
PREDICTED: transcription factor MUTE [Jatropha curcas] |
| 15 |
Hb_007441_090 |
0.2636250005 |
- |
- |
nucleoside diphosphate kinase 1 [Hevea brasiliensis] |
| 16 |
Hb_000029_100 |
0.2637806721 |
- |
- |
PREDICTED: nicalin-1-like isoform X2 [Populus euphratica] |
| 17 |
Hb_073799_010 |
0.2639253168 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_178620_010 |
0.2653743885 |
- |
- |
ADP-ribosylation factor, arf, putative [Ricinus communis] |
| 19 |
Hb_040747_010 |
0.2656000795 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 20 |
Hb_011287_040 |
0.2660176208 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810-like isoform X1 [Jatropha curcas] |