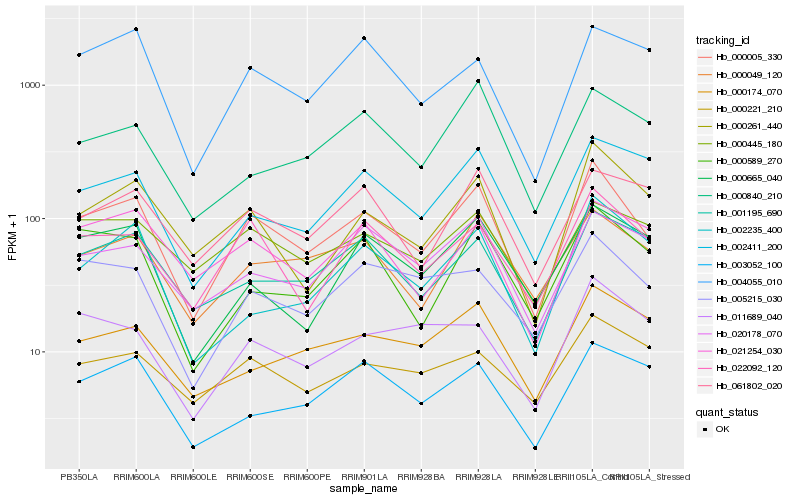
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000005_330 |
0.0 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 6 isoform X2 [Jatropha curcas] |
| 2 |
Hb_022092_120 |
0.1162569038 |
- |
- |
PREDICTED: uncharacterized protein LOC105646971 [Jatropha curcas] |
| 3 |
Hb_000049_120 |
0.1211740304 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 [Jatropha curcas] |
| 4 |
Hb_000261_440 |
0.1216218229 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_005215_030 |
0.1313942755 |
- |
- |
FAD-dependent oxidoreductase family protein [Populus trichocarpa] |
| 6 |
Hb_000589_270 |
0.1350098789 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Jatropha curcas] |
| 7 |
Hb_020178_070 |
0.1358095928 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 8 |
Hb_001195_690 |
0.1364763842 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185 [Jatropha curcas] |
| 9 |
Hb_002411_200 |
0.1367736786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_061802_020 |
0.1381381203 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 11 |
Hb_021254_030 |
0.1384972745 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein P [Jatropha curcas] |
| 12 |
Hb_000174_070 |
0.1398554879 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_000445_180 |
0.141488175 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At4g08455 [Jatropha curcas] |
| 14 |
Hb_000221_210 |
0.1417738459 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_004055_010 |
0.1429444719 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 16 |
Hb_003052_100 |
0.1437022694 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_002235_400 |
0.1439089595 |
- |
- |
PREDICTED: uncharacterized protein At5g19025-like isoform X1 [Populus euphratica] |
| 18 |
Hb_000665_040 |
0.1439983682 |
- |
- |
PREDICTED: stress response protein NST1-like [Jatropha curcas] |
| 19 |
Hb_000840_210 |
0.144549801 |
- |
- |
actin actin-like protein [Coniophora puteana RWD-64-598 SS2] |
| 20 |
Hb_011689_040 |
0.147073289 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |