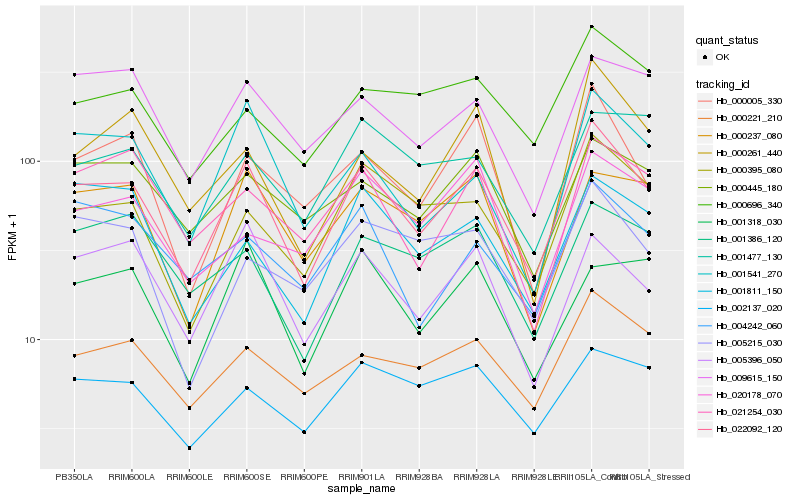
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022092_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646971 [Jatropha curcas] |
| 2 |
Hb_000221_210 |
0.1078984092 |
- |
- |
Kinase superfamily protein isoform 2 [Theobroma cacao] |
| 3 |
Hb_000395_080 |
0.112983524 |
- |
- |
PREDICTED: uncharacterized protein LOC105645573 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005396_050 |
0.1140785599 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase 4 [Jatropha curcas] |
| 5 |
Hb_000005_330 |
0.1162569038 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 6 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000237_080 |
0.1214952284 |
transcription factor |
TF Family: PHD |
PREDICTED: chromatin structure-remodeling complex subunit RSC1 [Jatropha curcas] |
| 7 |
Hb_002137_020 |
0.1215511432 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 8 |
Hb_001541_270 |
0.1219539417 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 9 |
Hb_000445_180 |
0.1228524811 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At4g08455 [Jatropha curcas] |
| 10 |
Hb_009615_150 |
0.1242252726 |
- |
- |
PREDICTED: transcription elongation factor SPT4 homolog 2 [Jatropha curcas] |
| 11 |
Hb_001386_120 |
0.1255243885 |
- |
- |
PREDICTED: yrdC domain-containing protein, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000696_340 |
0.1271637397 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 13 |
Hb_021254_030 |
0.1277008868 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein P [Jatropha curcas] |
| 14 |
Hb_000261_440 |
0.1310602683 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_004242_060 |
0.1316245205 |
- |
- |
PREDICTED: nicotinamidase 2-like [Jatropha curcas] |
| 16 |
Hb_005215_030 |
0.1322161313 |
- |
- |
FAD-dependent oxidoreductase family protein [Populus trichocarpa] |
| 17 |
Hb_001318_030 |
0.1329762389 |
- |
- |
PREDICTED: uncharacterized protein LOC105642686 [Jatropha curcas] |
| 18 |
Hb_001811_150 |
0.133585271 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_020178_070 |
0.1348860256 |
- |
- |
Protein Z, putative [Ricinus communis] |
| 20 |
Hb_001477_130 |
0.1350624095 |
- |
- |
PREDICTED: uncharacterized protein LOC105646871 [Jatropha curcas] |