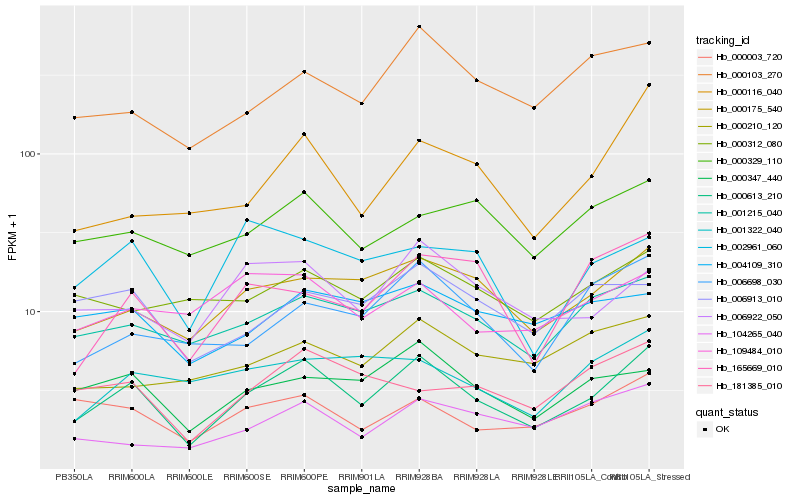
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000613_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000175_540 |
0.1394176842 |
- |
- |
PREDICTED: uncharacterized protein LOC105634886 [Jatropha curcas] |
| 3 |
Hb_001215_040 |
0.1498002462 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000347_440 |
0.1513432832 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 5 |
Hb_006922_050 |
0.1573300508 |
- |
- |
PREDICTED: dipeptidyl peptidase 8 [Jatropha curcas] |
| 6 |
Hb_000003_720 |
0.1589661778 |
- |
- |
hypothetical protein POPTR_0003s04720g [Populus trichocarpa] |
| 7 |
Hb_000103_270 |
0.1611188736 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_006698_030 |
0.162999553 |
- |
- |
PREDICTED: uncharacterized protein LOC105633969 [Jatropha curcas] |
| 9 |
Hb_181385_010 |
0.1650150978 |
- |
- |
PREDICTED: uncharacterized protein LOC105650319 [Jatropha curcas] |
| 10 |
Hb_000116_040 |
0.1680368874 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 11 |
Hb_165669_010 |
0.169219857 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_000329_110 |
0.1718858616 |
- |
- |
PREDICTED: uncharacterized protein LOC105643142 [Jatropha curcas] |
| 13 |
Hb_104265_040 |
0.1726599907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006913_010 |
0.1752322233 |
- |
- |
PREDICTED: uncharacterized protein LOC105649144 [Jatropha curcas] |
| 15 |
Hb_004109_310 |
0.1753227268 |
- |
- |
PREDICTED: uncharacterized protein LOC105646857 [Jatropha curcas] |
| 16 |
Hb_002961_060 |
0.1759842956 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 5 [Populus euphratica] |
| 17 |
Hb_000312_080 |
0.1763365741 |
- |
- |
PREDICTED: transmembrane protein 97-like [Jatropha curcas] |
| 18 |
Hb_001322_040 |
0.1774105475 |
- |
- |
- |
| 19 |
Hb_109484_010 |
0.1781242339 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 20 |
Hb_000210_120 |
0.1786409727 |
- |
- |
PREDICTED: uncharacterized protein LOC105635829 isoform X1 [Jatropha curcas] |