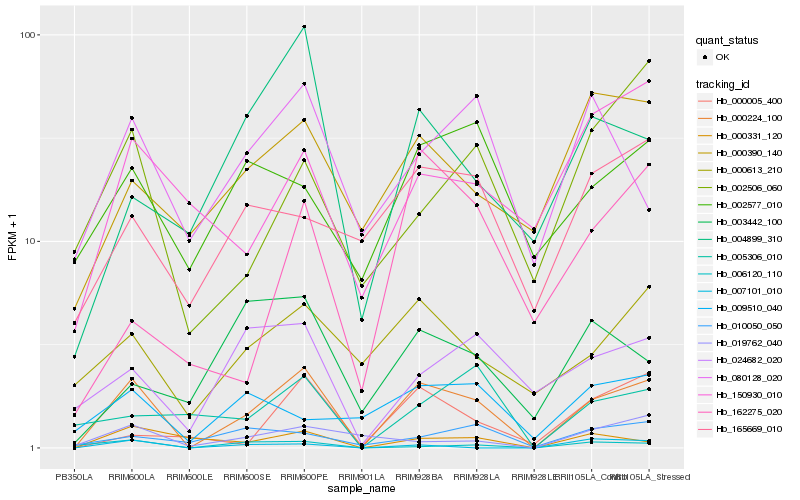
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000224_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_000331_120 |
0.2916583154 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006120_110 |
0.2988348041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000613_210 |
0.30327476 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 5 |
Hb_024682_020 |
0.3036336456 |
- |
- |
hypothetical protein POPTR_0001s12710g [Populus trichocarpa] |
| 6 |
Hb_000005_400 |
0.3057725042 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 23 isoform X2 [Jatropha curcas] |
| 7 |
Hb_004899_310 |
0.3084054465 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010050_050 |
0.313397535 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_005306_010 |
0.3182478437 |
- |
- |
- |
| 10 |
Hb_019762_040 |
0.3186125589 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000390_140 |
0.3212671743 |
- |
- |
PREDICTED: putative gamma-glutamylcyclotransferase At3g02910 [Jatropha curcas] |
| 12 |
Hb_007101_010 |
0.3218416512 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 13 |
Hb_009510_040 |
0.3248816762 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_165669_010 |
0.3255357148 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002577_010 |
0.3255558018 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 16 |
Hb_080128_020 |
0.3258174664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_162275_020 |
0.3261517661 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 18 |
Hb_003442_100 |
0.3266472092 |
- |
- |
PREDICTED: uncharacterized protein LOC105650128 [Jatropha curcas] |
| 19 |
Hb_002506_060 |
0.3273300142 |
- |
- |
PREDICTED: probable transporter mch1 [Populus euphratica] |
| 20 |
Hb_150930_010 |
0.3274952084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |