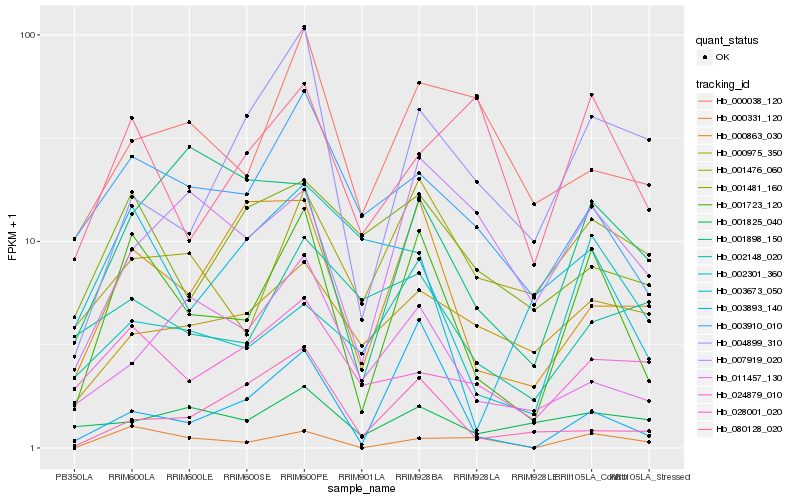
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001723_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003910_010 |
0.2522880138 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 3 |
Hb_000331_120 |
0.252723968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000863_030 |
0.2568699443 |
transcription factor |
TF Family: Orphans |
two-component system sensor histidine kinase/response regulator, putative [Ricinus communis] |
| 5 |
Hb_003893_140 |
0.2710984133 |
- |
- |
palmitoyl-acyl carrier protein thioesterase [Ricinus communis] |
| 6 |
Hb_001476_060 |
0.2743670032 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 7 |
Hb_002301_360 |
0.2811005598 |
- |
- |
hypothetical protein CICLE_v10017252mg [Citrus clementina] |
| 8 |
Hb_007919_020 |
0.2837893257 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 9 |
Hb_024879_010 |
0.2877452846 |
- |
- |
unknown [Glycine max] |
| 10 |
Hb_004899_310 |
0.2909750018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001898_150 |
0.2924008719 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 12 |
Hb_001481_160 |
0.2926398437 |
- |
- |
PREDICTED: RING-H2 finger protein ATL22-like [Populus euphratica] |
| 13 |
Hb_000038_120 |
0.2982148982 |
- |
- |
PREDICTED: endoglucanase 9-like [Jatropha curcas] |
| 14 |
Hb_080128_020 |
0.2987284677 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_011457_130 |
0.2997268023 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 16 |
Hb_001825_040 |
0.3023038328 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 17 |
Hb_028001_020 |
0.3024724282 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 18 |
Hb_003673_050 |
0.302794322 |
- |
- |
hypothetical protein POPTR_0001s40990g [Populus trichocarpa] |
| 19 |
Hb_002148_020 |
0.3059628597 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 20 |
Hb_000975_350 |
0.3079088972 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |