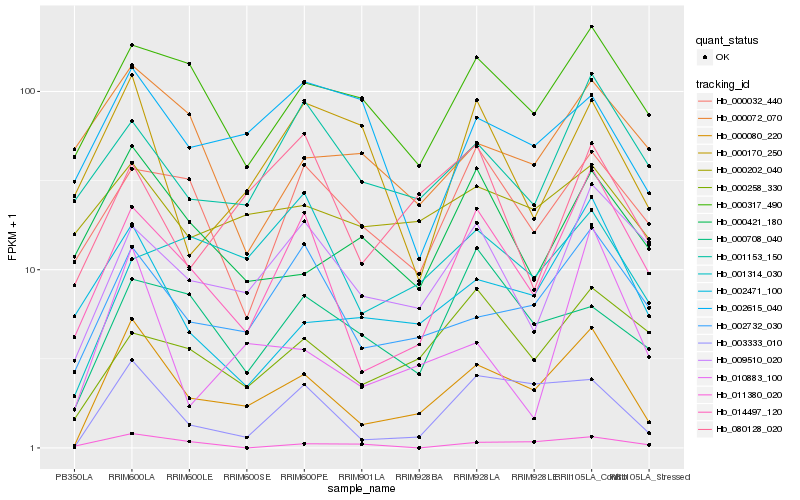
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000080_220 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_003333_010 |
0.1786025138 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 3 |
Hb_014497_120 |
0.1931317765 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 4 |
Hb_002732_030 |
0.2046931578 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002471_100 |
0.2295762378 |
- |
- |
PREDICTED: anthranilate synthase beta subunit 1, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000317_490 |
0.2328627646 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 7 |
Hb_009510_020 |
0.2345466727 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 8 |
Hb_010883_100 |
0.2411968711 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 9 |
Hb_000421_180 |
0.2440495416 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_001153_150 |
0.2494893572 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
HMG-CoA synthase 2 [Hevea brasiliensis] |
| 11 |
Hb_000202_040 |
0.2538246442 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BOI-like [Jatropha curcas] |
| 12 |
Hb_000258_330 |
0.2563509564 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 13 |
Hb_000032_440 |
0.2624420321 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_000708_040 |
0.2653144593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000170_250 |
0.2679398754 |
- |
- |
hypothetical protein F775_29828 [Aegilops tauschii] |
| 16 |
Hb_080128_020 |
0.2684221382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000072_070 |
0.2697876097 |
rubber biosynthesis |
Gene Name: Farnesyl diphosphate synthase |
farnesyl diphosphate synthase [Hevea brasiliensis] |
| 18 |
Hb_002615_040 |
0.2721985186 |
- |
- |
PREDICTED: uncharacterized protein LOC105110770 [Populus euphratica] |
| 19 |
Hb_001314_030 |
0.2747320754 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 20 |
Hb_011380_020 |
0.2752707056 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X4 [Populus euphratica] |