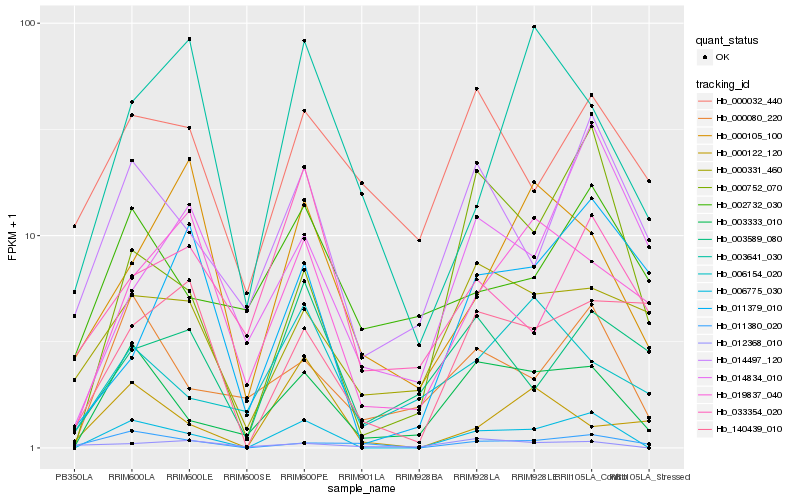
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006775_030 |
0.0 |
- |
- |
MADS-box transcription factor, partial [Clutia sp. DAV B80.252/F1980.8371] |
| 2 |
Hb_003333_010 |
0.2509475684 |
- |
- |
PREDICTED: uncharacterized protein LOC105645175 [Jatropha curcas] |
| 3 |
Hb_000752_070 |
0.2926671058 |
- |
- |
PREDICTED: mavicyanin-like [Jatropha curcas] |
| 4 |
Hb_000080_220 |
0.3016968202 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_014497_120 |
0.3082700315 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 6 |
Hb_011380_020 |
0.3227168949 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X4 [Populus euphratica] |
| 7 |
Hb_019837_040 |
0.3446813223 |
- |
- |
PREDICTED: histone-lysine N-methyltransferase SETD1B [Jatropha curcas] |
| 8 |
Hb_003589_080 |
0.3506459112 |
- |
- |
PREDICTED: uncharacterized protein LOC105107863 [Populus euphratica] |
| 9 |
Hb_012368_010 |
0.3547858332 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 10 |
Hb_002732_030 |
0.3583217348 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_014834_010 |
0.3612284947 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT4 [Jatropha curcas] |
| 12 |
Hb_003641_030 |
0.363333475 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 13 |
Hb_000331_460 |
0.3649584548 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 14 |
Hb_033354_020 |
0.3657331257 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_011379_010 |
0.3665894446 |
- |
- |
Thylakoid lumenal 25.6 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_006154_020 |
0.3672631306 |
transcription factor |
TF Family: C2H2 |
hypothetical protein RCOM_1621120 [Ricinus communis] |
| 17 |
Hb_000105_100 |
0.367543898 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_140439_010 |
0.3679369088 |
transcription factor |
TF Family: PLATZ |
protein with unknown function [Ricinus communis] |
| 19 |
Hb_000122_120 |
0.3695934089 |
- |
- |
PREDICTED: arogenate dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000032_440 |
0.370838457 |
- |
- |
protein with unknown function [Ricinus communis] |