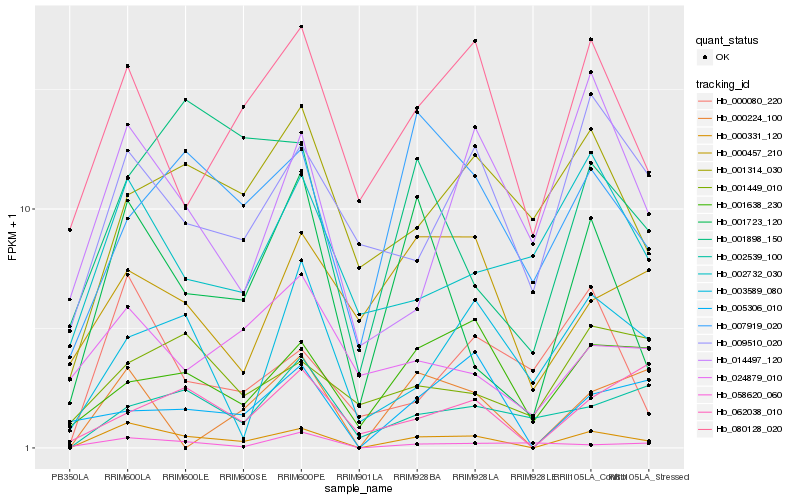
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001723_120 |
0.252723968 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_080128_020 |
0.2760414414 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009510_020 |
0.2908085128 |
- |
- |
PREDICTED: phosphatidylinositol transfer protein 2-like [Jatropha curcas] |
| 5 |
Hb_000224_100 |
0.2916583154 |
- |
- |
- |
| 6 |
Hb_000080_220 |
0.2923365589 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_007919_020 |
0.2971289071 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 8 |
Hb_001638_230 |
0.297861643 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 9 |
Hb_014497_120 |
0.2996226204 |
rubber biosynthesis |
Gene Name: Hydroxymethylglutaryl coenzyme A reductase |
RecName: Full=3-hydroxy-3-methylglutaryl-coenzyme A reductase 1; Short=HMG-CoA reductase 1 [Hevea brasiliensis] |
| 10 |
Hb_001314_030 |
0.3025521744 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 11 |
Hb_002732_030 |
0.3061745002 |
transcription factor |
TF Family: AP2 |
PREDICTED: ethylene-responsive transcription factor RAP2-7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001898_150 |
0.3069008313 |
- |
- |
PREDICTED: uncharacterized protein LOC105629671 [Jatropha curcas] |
| 13 |
Hb_002539_100 |
0.3072462829 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 14 |
Hb_003589_080 |
0.3093063495 |
- |
- |
PREDICTED: uncharacterized protein LOC105107863 [Populus euphratica] |
| 15 |
Hb_005306_010 |
0.3100776662 |
- |
- |
- |
| 16 |
Hb_058620_060 |
0.3115526817 |
- |
- |
uncharacterized protein LOC105646159 precursor [Jatropha curcas] |
| 17 |
Hb_024879_010 |
0.3129976648 |
- |
- |
unknown [Glycine max] |
| 18 |
Hb_062038_010 |
0.3130630105 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC104882266 isoform X2 [Vitis vinifera] |
| 19 |
Hb_000457_210 |
0.3192689512 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 20 |
Hb_001449_010 |
0.321655856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |