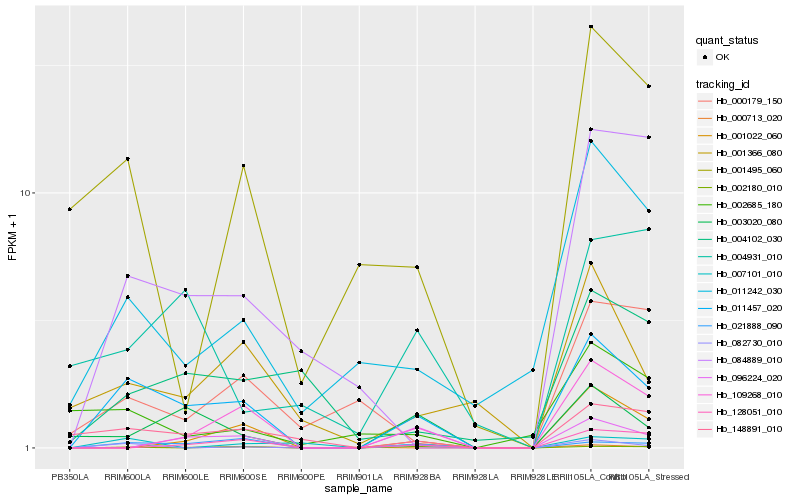
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011457_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649960 [Jatropha curcas] |
| 2 |
Hb_021888_090 |
0.2480854903 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 isoform X1 [Malus domestica] |
| 3 |
Hb_082730_010 |
0.266366693 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 4 |
Hb_148891_010 |
0.2873053639 |
- |
- |
- |
| 5 |
Hb_000713_020 |
0.3059771153 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 6 |
Hb_011242_030 |
0.3087212171 |
- |
- |
- |
| 7 |
Hb_003020_080 |
0.3107273589 |
- |
- |
- |
| 8 |
Hb_001366_080 |
0.3192786874 |
transcription factor |
TF Family: NAC |
NAC transcription factor 068 [Jatropha curcas] |
| 9 |
Hb_084889_010 |
0.3305323405 |
- |
- |
PREDICTED: uncharacterized protein LOC105638918 [Jatropha curcas] |
| 10 |
Hb_004102_030 |
0.3357731433 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 11 |
Hb_002180_010 |
0.3361883304 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_001495_060 |
0.3376128046 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 13 |
Hb_109268_010 |
0.3431758034 |
- |
- |
PREDICTED: uncharacterized protein LOC105650977 [Jatropha curcas] |
| 14 |
Hb_001022_060 |
0.3451739689 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 15 |
Hb_007101_010 |
0.3457669656 |
- |
- |
PREDICTED: 2-hydroxyisoflavanone dehydratase-like [Jatropha curcas] |
| 16 |
Hb_128051_010 |
0.3458216532 |
- |
- |
- |
| 17 |
Hb_004931_010 |
0.3515414164 |
- |
- |
inosine triphosphate pyrophosphatase, putative [Ricinus communis] |
| 18 |
Hb_096224_020 |
0.3589369508 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 19 |
Hb_000179_150 |
0.3593187302 |
- |
- |
hypothetical protein ZEAMMB73_153699 [Zea mays] |
| 20 |
Hb_002685_180 |
0.3604636939 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |