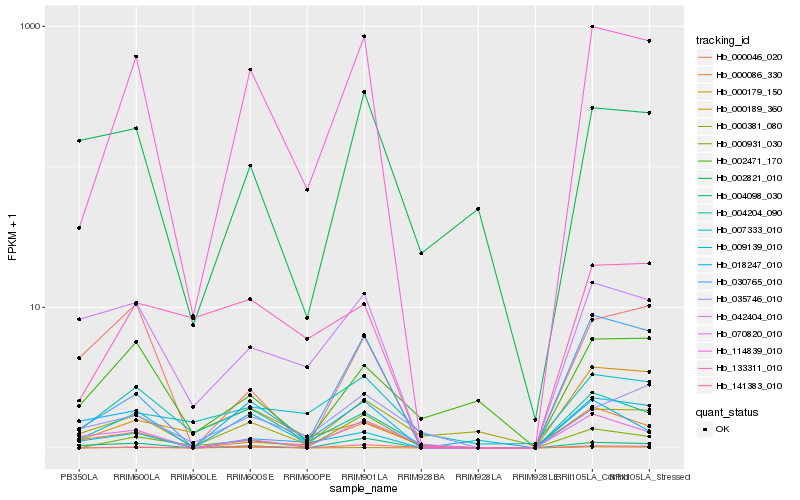
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114839_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000189_360 |
0.1942450438 |
- |
- |
Calcium-transporting ATPase 12, plasma membrane-type [Morus notabilis] |
| 3 |
Hb_000086_330 |
0.2291920009 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 4 |
Hb_000931_030 |
0.2396378212 |
- |
- |
PREDICTED: uncharacterized protein LOC105130278 [Populus euphratica] |
| 5 |
Hb_042404_010 |
0.2408501982 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 6 |
Hb_004098_030 |
0.2412868868 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 7 |
Hb_070820_010 |
0.2427396409 |
- |
- |
hypothetical protein B456_008G232800 [Gossypium raimondii] |
| 8 |
Hb_004204_090 |
0.2487559077 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |
| 9 |
Hb_035746_010 |
0.2506445884 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 10 |
Hb_133311_010 |
0.2526531778 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like isoform X1 [Glycine max] |
| 11 |
Hb_141383_010 |
0.2578105469 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 12 |
Hb_007333_010 |
0.2607262392 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_009139_010 |
0.2665881959 |
- |
- |
putative Phospholipid-Diacylglycerol acyltransferase [Elaeis guineensis] |
| 14 |
Hb_000179_150 |
0.269457469 |
- |
- |
hypothetical protein ZEAMMB73_153699 [Zea mays] |
| 15 |
Hb_018247_010 |
0.2720415915 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 16 |
Hb_000046_020 |
0.2732215771 |
- |
- |
- |
| 17 |
Hb_000381_080 |
0.2756085078 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair protein RAD16 [Jatropha curcas] |
| 18 |
Hb_002471_170 |
0.275700692 |
- |
- |
- |
| 19 |
Hb_030765_010 |
0.2770632525 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter G family member 11-like isoform X1 [Solanum lycopersicum] |
| 20 |
Hb_002821_010 |
0.2777025179 |
- |
- |
PREDICTED: nucleolar protein 16 [Jatropha curcas] |